Conservation of protein kinase a catalytic subunit sequences in the schistosome pathogens of humans
- PMID: 20109453
- PMCID: PMC2859104
- DOI: 10.1016/j.exppara.2010.01.012
Conservation of protein kinase a catalytic subunit sequences in the schistosome pathogens of humans
Abstract
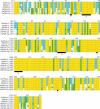
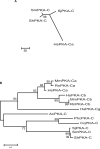
cAMP-dependent protein kinases (PKAs) are central mediators of cAMP signaling in eukaryotic cells. Previously we identified a cDNA which encodes for a PKA catalytic subunit (PKA-C) in Schistosoma mansoni (SmPKA-C) that is required for adult schistosome viability in vitro. As such, SmPKA-C could potentially represent a novel schistosome chemotherapeutic target. Here we sought to identify PKA-C subunit orthologues in the other medically important schistosome species, Schistosoma haematobium and Schistosoma japonicum, to determine the degree to which this potential target is conserved and could therefore be exploited for the treatment of all forms of schistosomiasis. We report the identification of PKA-C subunit orthologues in S. haematobium and S. japonicum (ShPKA-C and SjPKA-C, respectively) and show that PKA-C orthologues are highly conserved in the Schistosoma, with over 99% amino acid sequence identity shared among the three human pathogens we examined. Furthermore, we show that the recently published Schistosoma mansoni and S. japonicum genomes contain sequences encoding for several putative PKA substrates with homology to those found in Homo sapiens, Caenorhabditis elegans, and Saccharomyces cerevisiae.
Figures
Similar articles
-
Lysophospholipase from the human blood fluke, Schistosoma japonicum.Int J Infect Dis. 2008 Mar;12(2):143-51. doi: 10.1016/j.ijid.2007.05.005. Epub 2007 Aug 20. Int J Infect Dis. 2008. PMID: 17709268
-
Mapping and sequencing of acetylcholinesterase genes from the platyhelminth blood fluke Schistosoma.Gene. 2003 Sep 18;314:103-12. doi: 10.1016/s0378-1119(03)00709-1. Gene. 2003. PMID: 14527722
-
Schistosoma japonicum: analysis of eggshell protein genes, their expression, and comparison with similar genes from other schistosomes.Exp Parasitol. 1991 May;72(4):381-90. doi: 10.1016/0014-4894(91)90084-a. Exp Parasitol. 1991. PMID: 1709112
-
Gaining biological perspectives from schistosome genomes.Mol Biochem Parasitol. 2014 Aug;196(1):21-8. doi: 10.1016/j.molbiopara.2014.07.007. Epub 2014 Jul 27. Mol Biochem Parasitol. 2014. PMID: 25076011 Review.
-
Molecular cloning of schistosome genes.Parasitology. 1986;92 Suppl:S73-81. doi: 10.1017/s003118200008570x. Parasitology. 1986. PMID: 2940502 Review. No abstract available.
Cited by
-
Defining the Schistosoma haematobium kinome enables the prediction of essential kinases as anti-schistosome drug targets.Sci Rep. 2015 Dec 4;5:17759. doi: 10.1038/srep17759. Sci Rep. 2015. PMID: 26635209 Free PMC article.
References
-
- Caffrey CR. Chemotherapy of schistosomiasis: present and future. Current Opinion in Chemical Biology. 2007;11:433–439. - PubMed
-
- Cioli D, Pica-Mattoccia L. Praziquantel. Parasitology Research. 2003;90(Supp 1):S3–9. - PubMed
-
- Dissous C, Ahier A, Khayath N. Protein tyrosine kinases as new potential targets against human schistosomiasis. Bioessays. 2007;29:1281–1288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

