Genomic characterization of human adenovirus 36, a putative obesity agent
- PMID: 20109503
- PMCID: PMC7021214
- DOI: 10.1016/j.virusres.2010.01.011
Genomic characterization of human adenovirus 36, a putative obesity agent
Abstract
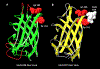
Increased levels of serum antibody titers against human adenovirus 36 (HAdV-D36) are associated with human obesity and experimental obesity in laboratory animals. While HAdV-D36 has been studied as an infectious agent implicated in obesity for over a decade, the complete genome sequence and its analysis have yet to be reported. A detailed analysis of the genome sequence of HAdV-D36 may be important to understand its role in obesity. Genomic and bioinformatic comparisons with other HAdVs identified differences that suggested unique functions. Global pairwise genome alignment with all sequenced human adenovirus D (HAdV-D) genomes revealed areas of nonconserved sequences in the hexon, E3 CR1 beta, E3 CR1 gamma, and fiber genes. Phylogenetic analysis of all HAdV-D36 proteins confirmed that this virus belongs to species Human adenovirus D. This genomic analysis of HAdV-D36 provides an important tool for comprehending the role that this unique adenovirus may play in human obesity. Low amino acid sequence identity in the E3 CR1 beta and CR1 gamma genes may suggest distinctive roles for these proteins. Furthermore, the predicted molecular models of the HAdV-D36 fiber protein seem to implicate a unique tissue tropism for HAdV-D36.
(c) 2010 Elsevier B.V. All rights reserved.
Figures




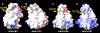
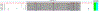

References
-
- Atkinson RL, Dhurandhar NV, Allison DB, Bowen RL, Israel BA, Albu JB, Augustus AS, 2005. Human adenovirus-36 is associated with increased body weight and paradoxical reduction of serum lipids. Int. J. Obes. (Lond.) 29 (3), 281–286. - PubMed
-
- Brown WV, Fujioka K, Wilson PW, Woodworth KA, 2009. Obesity: why be concerned? Am. J. Med 122 (4 (Suppl. 1)), S4–S11. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

