NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA
- PMID: 20110252
- PMCID: PMC2879506
- DOI: 10.1093/nar/gkq020
NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA
Abstract
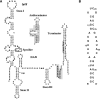
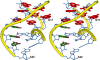
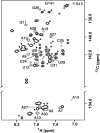
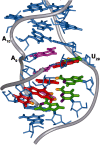
Gram-positive bacteria utilize a tRNA-responsive transcription antitermination mechanism, designated the T box system, to regulate expression of many amino acid biosynthetic and aminoacyl-tRNA synthetase genes. The RNA transcripts of genes controlled by this mechanism contain 5' untranslated regions, or leader RNAs, that specifically bind cognate tRNA molecules through pairing of nucleotides in the tRNA anticodon loop with nucleotides in the Specifier Loop domain of the leader RNA. We have determined the solution structure of the Specifier Loop domain of the tyrS leader RNA from Bacillus subtilis. Fifty percent of the nucleotides in the Specifier Loop domain adopt a loop E motif. The Specifier Sequence nucleotides, which pair with the tRNA anticodon, stack with their Watson-Crick edges rotated toward the minor groove and exhibit only modest flexibility. We also show that a Specifier Loop domain mutation that impairs the function of the B. subtilis glyQS T box RNA disrupts the tyrS loop E motif. Our results suggest a mechanism for tRNA-Specifier Loop binding in which the phosphate backbone kink created by the loop E motif causes the Specifier Sequence bases to rotate toward the minor groove, which increases accessibility for pairing with bases in the anticodon loop of tRNA.
Figures

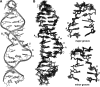
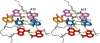
Similar articles
-
Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA.J Mol Biol. 2011 Apr 22;408(1):99-117. doi: 10.1016/j.jmb.2011.02.014. Epub 2011 Feb 17. J Mol Biol. 2011. PMID: 21333656 Free PMC article.
-
Specificity of tRNA-mRNA interactions in Bacillus subtilis tyrS antitermination.J Bacteriol. 1997 Apr;179(8):2587-94. doi: 10.1128/jb.179.8.2587-2594.1997. J Bacteriol. 1997. PMID: 9098057 Free PMC article.
-
Solution NMR determination of hydrogen bonding and base pairing between the glyQS T box riboswitch Specifier domain and the anticodon loop of tRNA(Gly).FEBS Lett. 2013 Nov 1;587(21):3495-9. doi: 10.1016/j.febslet.2013.09.003. Epub 2013 Sep 11. FEBS Lett. 2013. PMID: 24036450 Free PMC article.
-
An evolving tale of two interacting RNAs-themes and variations of the T-box riboswitch mechanism.IUBMB Life. 2019 Aug;71(8):1167-1180. doi: 10.1002/iub.2098. Epub 2019 Jun 17. IUBMB Life. 2019. PMID: 31206978 Free PMC article. Review.
-
Aminoacyl-tRNA synthetase genes of Bacillus subtilis: organization and regulation.Biochem Cell Biol. 1999;77(4):343-7. Biochem Cell Biol. 1999. PMID: 10546897 Review.
Cited by
-
The T box riboswitch: A novel regulatory RNA that utilizes tRNA as its ligand.Biochim Biophys Acta. 2014 Oct;1839(10):959-963. doi: 10.1016/j.bbagrm.2014.04.022. Epub 2014 May 9. Biochim Biophys Acta. 2014. PMID: 24816551 Free PMC article. Review.
-
Sequence-based identification of 3D structural modules in RNA with RMDetect.Nat Methods. 2011 Jun;8(6):513-21. doi: 10.1038/nmeth.1603. Epub 2011 May 8. Nat Methods. 2011. PMID: 21552257
-
Targeting Gene Transcription Prevents Antibiotic Resistance.Antibiotics (Basel). 2025 Mar 27;14(4):345. doi: 10.3390/antibiotics14040345. Antibiotics (Basel). 2025. PMID: 40298535 Free PMC article. Review.
-
Two-codon T-box riboswitch binding two tRNAs.Proc Natl Acad Sci U S A. 2013 Jul 30;110(31):12756-61. doi: 10.1073/pnas.1304307110. Epub 2013 Jul 15. Proc Natl Acad Sci U S A. 2013. PMID: 23858450 Free PMC article.
-
Global analysis of riboswitches by small-angle X-ray scattering and calorimetry.Biochim Biophys Acta. 2014 Oct;1839(10):1020-1029. doi: 10.1016/j.bbagrm.2014.04.014. Epub 2014 Apr 24. Biochim Biophys Acta. 2014. PMID: 24769285 Free PMC article. Review.
References
-
- Henkin TM, Grundy FJ. Sensing metabolic signals with nascent RNA transcripts: the T box and S box riboswitches as paradigms. Cold Spring Harbor Symp. Quant. Biol. 2006;71:231–237. - PubMed
-
- Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

