Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX
- PMID: 20110259
- PMCID: PMC2875037
- DOI: 10.1093/nar/gkq006
Involvement of histone deacetylation in MORC2-mediated down-regulation of carbonic anhydrase IX
Abstract
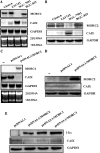
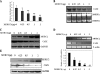
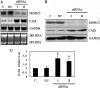
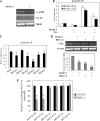
Carbonic anhydrase IX (CAIX) plays an important role in the growth and survival of tumor cells. MORC2 is a member of the MORC protein family. The MORC proteins contain a CW-type zinc finger domain and are predicted to have the function of regulating transcription, but no MORC2 target genes have been identified. Here we performed a DNA microarray hybridization and found CAIX mRNA to be down-regulated 8-fold when MORC2 was overexpressed. This result was further confirmed by northern and western blot analysis. Our results also showed that the protected region 4 (PR4) was important for the repression function of MORC2. Moreover, MORC2 decreased the acetylation level of histone H3 at the CAIX promoter. Meanwhile, trichostatin A (TSA) had an increasing effect on CAIX promoter activity. Among the six HDACs tested, histone deacetylase 4 (HDAC4) had a much more prominent effect on CAIX repression. ChIP and ChIP Re-IP assays showed that MORC2 and HDAC4 were assembled on the same region of the CAIX promoter. Importantly, we further confirmed that both proteins are simultaneously present in the PR4-binding complex. These results may contribute to understanding the molecular mechanisms of CAIX regulation.
Figures



References
-
- Robertson N, Potter C, Harris AL. Role of carbonic anhydrase IX in human tumor cell growth, survival and invasion. Cancer Res. 2004;64:6160–6165. - PubMed
-
- Potter C, Harris AL. Hypoxia inducible carbonic anhydrase IX, marker of tumor hypoxia, survival pathway and therapy target. Cell Cycle. 2004;3:164–167. - PubMed
-
- Gut MO, Parkkila S, Vernerová Z, Rohde E, Závada J, Höcker M, Pastorek J, Karttunen T, Gibadulinová A, Závadová Z, et al. Gastric hyperplasia in mice with targeted disruption of the carbonic anhydrase gene Car9. Gastroenterology. 2002;123:1889–1903. - PubMed
-
- Driessen A, Landuyt W, Pastorekova S, Moons J, Goethals L, Haustermans K, Nafteux P, Penninckx F, Geboes K, Lerut T, et al. Expression of Carbonic Anhydrase IX (CA IX), a hypoxia-related protein, rather than Vascular-Endothelial Growth Factor (VEGF), a pro-angiogenic factor, correlates with an extremely poor prognosis in esophageal and gastric adenocarcinomas. Ann. Surg. 2006;243:334–340. - PMC - PubMed
-
- Airley RE, Loncaster J, Raleigh JA, Harris AL, Davidson SE, Hunter RD, West CM, Stratford IJ. GLUT-1 and CAIX as intrinsic markers of hypoxia in carcinoma of the cervix: relationship to pimonidazole binding. Int. J. Cancer. 2003;104:85–91. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

