Evolutionary optimization of a modular ligase ribozyme: a small catalytic unit and a hairpin motif masking an element that could form an inactive structure
- PMID: 20110262
- PMCID: PMC2879505
- DOI: 10.1093/nar/gkq018
Evolutionary optimization of a modular ligase ribozyme: a small catalytic unit and a hairpin motif masking an element that could form an inactive structure
Abstract
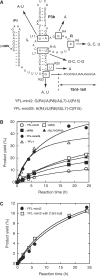
The YFL ribozyme is an artificial ligase ribozyme isolated by a 'design and selection' strategy, in which a modular catalytic unit was generated on a rationally designed modular scaffold RNA. This ligase ribozyme has a versatile catalytic unit that accepts not only beta-nicotinamide mononucleotide (beta-NMN) but also inorganic pyrophosphate as leaving groups for template-dependent RNA ligation. Although this property is interesting from an evolutionary viewpoint regarding primitive RNA ligation/polymerization systems in the RNA world, structural analysis of the YFL ribozyme has not been continued due to apparent structural nonuniformity of its folded state. To elucidate the active structure of the YFL ribozyme, we performed in vitro evolution experiments to improve its folding ability. Biochemical and phylogenetic analyses of evolved variants indicated that the catalytic unit of the YFL ribozyme is compact and the 3' single-stranded region of the parent YFL-1 ribozyme contributes to mask an element that could form an inactive structure.
Figures





Similar articles
-
Generation and development of RNA ligase ribozymes with modular architecture through "design and selection".Molecules. 2010 Aug 26;15(9):5850-65. doi: 10.3390/molecules15095850. Molecules. 2010. PMID: 22273983 Free PMC article. Review.
-
Tailoring RNA modular units on a common scaffold: a modular ribozyme with a catalytic unit for beta-nicotinamide mononucleotide-activated RNA ligation.RNA. 2009 May;15(5):877-88. doi: 10.1261/rna.1461309. Epub 2009 Mar 23. RNA. 2009. PMID: 19307294 Free PMC article.
-
Construction of an artificial ribozyme which ligates an RNA fragment activated by nicotinamide mononucleotide.Nucleic Acids Symp Ser (Oxf). 2006;(50):231-2. doi: 10.1093/nass/nrl115. Nucleic Acids Symp Ser (Oxf). 2006. PMID: 17150902
-
RNA-catalyzed RNA ligation on an external RNA template.Chem Biol. 2002 Mar;9(3):297-307. doi: 10.1016/s1074-5521(02)00110-2. Chem Biol. 2002. PMID: 11927255
-
Structure and function of the hairpin ribozyme.J Mol Biol. 2000 Mar 24;297(2):269-91. doi: 10.1006/jmbi.2000.3560. J Mol Biol. 2000. PMID: 10715200 Review.
Cited by
-
Deep sequencing analysis of mutations resulting from the incorporation of dNTP analogs.Nucleic Acids Res. 2010 Dec;38(22):8095-104. doi: 10.1093/nar/gkq685. Epub 2010 Aug 6. Nucleic Acids Res. 2010. PMID: 20693528 Free PMC article.
-
Generation and development of RNA ligase ribozymes with modular architecture through "design and selection".Molecules. 2010 Aug 26;15(9):5850-65. doi: 10.3390/molecules15095850. Molecules. 2010. PMID: 22273983 Free PMC article. Review.
-
A range of complex probabilistic models for RNA secondary structure prediction that includes the nearest-neighbor model and more.RNA. 2012 Feb;18(2):193-212. doi: 10.1261/rna.030049.111. Epub 2011 Dec 22. RNA. 2012. PMID: 22194308 Free PMC article.
References
-
- Campbell ID, Baron M. The structure and function of protein modules. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1991;332:165–170. - PubMed
-
- Campbell ID, Downing AK. Building protein structure and function from modular units. Trends Biotechnol. 1994;12:168–172. - PubMed
-
- Brion P, Westhof E. Hierarchy and dynamics of RNA folding. Annu. Rev. Biophys. Biomol. Struct. 1997;26:113–137. - PubMed
-
- Jaeger L. The new world of ribozymes. Curr. Opin. Struct. Biol. 1997;7:324–335. - PubMed
-
- Westhof E, Masquida B, Jaeger L. RNA tectonics: towards RNA design. Fold. Des. 1996;1:R78–R88. - PubMed

