Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks
- PMID: 20110345
- PMCID: PMC2836973
- DOI: 10.1091/mbc.e09-11-0958
Rad17 plays a central role in establishment of the interaction between TopBP1 and the Rad9-Hus1-Rad1 complex at stalled replication forks
Abstract
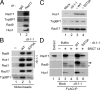
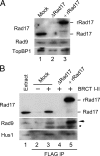
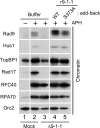
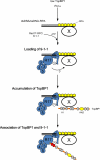
Rad17 is critical for the ATR-dependent activation of Chk1 during checkpoint responses. It is known that Rad17 loads the Rad9-Hus1-Rad1 (9-1-1) complex onto DNA. We show that Rad17 also mediates the interaction of 9-1-1 with the ATR-activating protein TopBP1 in Xenopus egg extracts. Studies with Rad17 mutants indicate that binding of ATP to Rad17 is essential for the association of 9-1-1 and TopBP1. Furthermore, hydrolysis of ATP by Rad17 is necessary for the loading of 9-1-1 onto DNA and the elevated, checkpoint-dependent accumulation of TopBP1 on chromatin. Significantly, a mutant 9-1-1 complex that cannot bind TopBP1 has a normal capacity to promote elevated accumulation of TopBP1 on chromatin. Taken together, we propose the following mechanism. First, Rad17 loads 9-1-1 onto DNA. Second, TopBP1 accumulates on chromatin in a manner that depends on both Rad17 and 9-1-1. Finally, 9-1-1 and TopBP1 dock in a Rad17-dependent manner before activation of Chk1.
Figures


Similar articles
-
TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks.J Cell Biol. 2009 Mar 23;184(6):793-804. doi: 10.1083/jcb.200810185. Epub 2009 Mar 16. J Cell Biol. 2009. PMID: 19289795 Free PMC article.
-
The Mre11-Rad50-Nbs1 (MRN) complex has a specific role in the activation of Chk1 in response to stalled replication forks.Mol Biol Cell. 2013 May;24(9):1343-53. doi: 10.1091/mbc.E13-01-0025. Epub 2013 Mar 6. Mol Biol Cell. 2013. PMID: 23468519 Free PMC article.
-
Interaction between Rad9-Hus1-Rad1 and TopBP1 activates ATR-ATRIP and promotes TopBP1 recruitment to sites of UV-damage.DNA Repair (Amst). 2014 Sep;21:1-11. doi: 10.1016/j.dnarep.2014.05.001. Epub 2014 May 27. DNA Repair (Amst). 2014. PMID: 25091155
-
TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: implications for a replication restart-based mechanism for ATR checkpoint activation.Cell Cycle. 2009 Sep 15;8(18):2877-84. doi: 10.4161/cc.8.18.9485. Epub 2009 Sep 9. Cell Cycle. 2009. PMID: 19652550 Review.
-
Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex.DNA Repair (Amst). 2004 Aug-Sep;3(8-9):1009-14. doi: 10.1016/j.dnarep.2004.03.032. DNA Repair (Amst). 2004. PMID: 15279787 Review.
Cited by
-
Critical role of SMG7 in activation of the ATR-CHK1 axis in response to genotoxic stress.Sci Rep. 2021 Apr 5;11(1):7502. doi: 10.1038/s41598-021-86957-x. Sci Rep. 2021. PMID: 33820915 Free PMC article.
-
CtIP interacts with TopBP1 and Nbs1 in the response to double-stranded DNA breaks (DSBs) in Xenopus egg extracts.Cell Cycle. 2011 Feb 1;10(3):469-80. doi: 10.4161/cc.10.3.14711. Epub 2011 Feb 1. Cell Cycle. 2011. PMID: 21263215 Free PMC article.
-
ATR: a master conductor of cellular responses to DNA replication stress.Trends Biochem Sci. 2011 Mar;36(3):133-40. doi: 10.1016/j.tibs.2010.09.005. Epub 2010 Oct 12. Trends Biochem Sci. 2011. PMID: 20947357 Free PMC article. Review.
-
A scalable platform for efficient CRISPR-Cas9 chemical-genetic screens of DNA damage-inducing compounds.Sci Rep. 2024 Jan 30;14(1):2508. doi: 10.1038/s41598-024-51735-y. Sci Rep. 2024. PMID: 38291084 Free PMC article.
-
NONO regulates the intra-S-phase checkpoint in response to UV radiation.Oncogene. 2016 Feb 4;35(5):567-76. doi: 10.1038/onc.2015.107. Epub 2015 Apr 20. Oncogene. 2016. PMID: 25893301
References
-
- Bowman G. D., O'Donnell M., Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–730. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

