Identification and characterization of gamma-glutamylamine cyclotransferase, an enzyme responsible for gamma-glutamyl-epsilon-lysine catabolism
- PMID: 20110353
- PMCID: PMC2843214
- DOI: 10.1074/jbc.M109.082099
Identification and characterization of gamma-glutamylamine cyclotransferase, an enzyme responsible for gamma-glutamyl-epsilon-lysine catabolism
Abstract
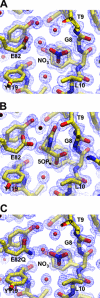
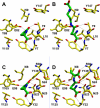
Gamma-glutamylamine cyclotransferase (GGACT) is an enzyme that converts gamma-glutamylamines to free amines and 5-oxoproline. GGACT shows high activity toward gamma-glutamyl-epsilon-lysine, derived from the breakdown of fibrin and other proteins cross-linked by transglutaminases. The enzyme adopts the newly identified cyclotransferase fold, observed in gamma-glutamylcyclotransferase (GGCT), an enzyme with activity toward gamma-glutamyl-alpha-amino acids (Oakley, A. J., Yamada, T., Liu, D., Coggan, M., Clark, A. G., and Board, P. G. (2008) J. Biol. Chem. 283, 22031-22042). Despite the absence of significant sequence identity, several residues are conserved in the active sites of GGCT and GGACT, including a putative catalytic acid/base residue (GGACT Glu(82)). The structure of GGACT in complex with the reaction product 5-oxoproline provides evidence for a common catalytic mechanism in both enzymes. The proposed mechanism, combined with the three-dimensional structures, also explains the different substrate specificities of these enzymes. Despite significant sequence divergence, there are at least three subfamilies in prokaryotes and eukaryotes that have conserved the GGCT fold and GGCT enzymatic activity.
Figures
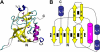


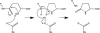
Similar articles
-
Probing the specificity of gamma-glutamylamine cyclotransferase: an enzyme involved in the metabolism of transglutaminase-catalyzed protein crosslinks.Amino Acids. 2013 Jan;44(1):143-50. doi: 10.1007/s00726-011-1153-2. Epub 2011 Nov 27. Amino Acids. 2013. PMID: 22120669
-
The identification and structural characterization of C7orf24 as gamma-glutamyl cyclotransferase. An essential enzyme in the gamma-glutamyl cycle.J Biol Chem. 2008 Aug 8;283(32):22031-42. doi: 10.1074/jbc.M803623200. Epub 2008 May 30. J Biol Chem. 2008. PMID: 18515354
-
gamma-Glutamylamine cyclotransferase. An enzyme involved in the catabolism of epsilon-(gamma-glutamyl)lysine and other gamma-glutamylamines.Mol Cell Biochem. 1981 Aug 11;38 Spec No(Pt 1):59-67. doi: 10.1007/BF00235688. Mol Cell Biochem. 1981. PMID: 6117008
-
γ-Glutamylamines and neurodegenerative diseases.Amino Acids. 2013 Jan;44(1):129-42. doi: 10.1007/s00726-011-1209-3. Epub 2012 Mar 10. Amino Acids. 2013. PMID: 22407484 Free PMC article. Review.
-
Mechanisms of Tumor Growth Inhibition by Depletion of γ-Glutamylcyclotransferase (GGCT): A Novel Molecular Target for Anticancer Therapy.Int J Mol Sci. 2018 Jul 14;19(7):2054. doi: 10.3390/ijms19072054. Int J Mol Sci. 2018. PMID: 30011933 Free PMC article. Review.
Cited by
-
Transglutaminse 2 and EGGL, the protein cross-link formed by transglutaminse 2, as therapeutic targets for disabilities of old age.Rejuvenation Res. 2013 Dec;16(6):495-517. doi: 10.1089/rej.2013.1452. Rejuvenation Res. 2013. PMID: 23968147 Free PMC article. Review.
-
Helicobacter pylori induces somatic mutations in TP53 via overexpression of CHAC1 in infected gastric epithelial cells.FEBS Open Bio. 2018 Mar 9;8(4):671-679. doi: 10.1002/2211-5463.12402. eCollection 2018 Apr. FEBS Open Bio. 2018. PMID: 29632819 Free PMC article.
-
Emerging regulatory paradigms in glutathione metabolism.Adv Cancer Res. 2014;122:69-101. doi: 10.1016/B978-0-12-420117-0.00002-5. Adv Cancer Res. 2014. PMID: 24974179 Free PMC article. Review.
-
ChaC2, an Enzyme for Slow Turnover of Cytosolic Glutathione.J Biol Chem. 2017 Jan 13;292(2):638-651. doi: 10.1074/jbc.M116.727479. Epub 2016 Dec 2. J Biol Chem. 2017. PMID: 27913623 Free PMC article.
-
Tn-Seq reveals hidden complexity in the utilization of host-derived glutathione in Francisella tularensis.PLoS Pathog. 2020 Jun 3;16(6):e1008566. doi: 10.1371/journal.ppat.1008566. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32492066 Free PMC article.
References
-
- Pisano J. J., Finlayson J. S., Peyton M. P. (1968) Science 160, 892–893 - PubMed
-
- Fink M. L., Folk J. E. (1981) Mol. Cell. Biochem. 38, 59–67 - PubMed
-
- Oakley A. J., Yamada T., Liu D., Coggan M., Clark A. G., Board P. G. (2008) J. Biol. Chem. 283, 22031–22042 - PubMed
-
- Orlowski M., Richman P. G., Meister A. (1969) Biochemistry 8, 1048–1055 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

