Structure and function of factor XI
- PMID: 20110423
- PMCID: PMC4828079
- DOI: 10.1182/blood-2009-09-199182
Structure and function of factor XI
Abstract
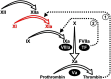
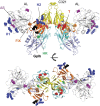
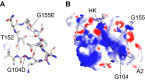
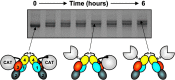
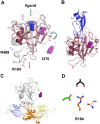
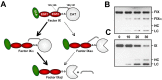
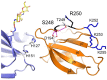
Factor XI (FXI) is the zymogen of an enzyme (FXIa) that contributes to hemostasis by activating factor IX. Although bleeding associated with FXI deficiency is relatively mild, there has been resurgence of interest in FXI because of studies indicating it makes contributions to thrombosis and other processes associated with dysregulated coagulation. FXI is an unusual dimeric protease, with structural features that distinguish it from vitamin K-dependent coagulation proteases. The recent availability of crystal structures for zymogen FXI and the FXIa catalytic domain have enhanced our understanding of structure-function relationships for this molecule. FXI contains 4 "apple domains" that form a disk structure with extensive interfaces at the base of the catalytic domain. The characterization of the apple disk structure, and its relationship to the catalytic domain, have provided new insight into the mechanism of FXI activation, the interaction of FXIa with the substrate factor IX, and the binding of FXI to platelets. Analyses of missense mutations associated with FXI deficiency have provided additional clues to localization of ligand-binding sites on the protein surface. Together, these data will facilitate efforts to understand the physiology and pathology of this unusual protease, and development of therapeutics to treat thrombotic disorders.
Figures

Similar articles
-
Model for a factor IX activation complex on blood platelets: dimeric conformation of factor XIa is essential.Blood. 2001 May 15;97(10):3117-22. doi: 10.1182/blood.v97.10.3117. Blood. 2001. PMID: 11342438
-
Analysis of the factor XI variant Arg184Gly suggests a structural basis for factor IX binding to factor XIa.J Thromb Haemost. 2013 Jul;11(7):1374-84. doi: 10.1111/jth.12275. J Thromb Haemost. 2013. PMID: 23617568 Free PMC article.
-
Hydrogen-deuterium exchange mass spectrometry highlights conformational changes induced by factor XI activation and binding of factor IX to factor XIa.J Thromb Haemost. 2019 Dec;17(12):2047-2055. doi: 10.1111/jth.14632. Epub 2019 Oct 7. J Thromb Haemost. 2019. PMID: 31519061 Free PMC article.
-
Structural and functional features of factor XI.J Thromb Haemost. 2009 Jul;7 Suppl 1(Suppl 1):75-8. doi: 10.1111/j.1538-7836.2009.03414.x. J Thromb Haemost. 2009. PMID: 19630773 Free PMC article. Review.
-
The mechanism underlying activation of factor IX by factor XIa.Thromb Res. 2014 May;133 Suppl 1(0 1):S48-51. doi: 10.1016/j.thromres.2014.03.020. Thromb Res. 2014. PMID: 24759143 Free PMC article. Review.
Cited by
-
Factor XI Deficiency Alters the Cytokine Response and Activation of Contact Proteases during Polymicrobial Sepsis in Mice.PLoS One. 2016 Apr 5;11(4):e0152968. doi: 10.1371/journal.pone.0152968. eCollection 2016. PLoS One. 2016. PMID: 27046148 Free PMC article.
-
DAKS1, a Kunitz Scaffold Peptide from the Venom Gland of Deinagkistrodon acutus Prevents Carotid-Artery and Middle-Cerebral-Artery Thrombosis via Targeting Factor XIa.Pharmaceuticals (Basel). 2021 Sep 24;14(10):966. doi: 10.3390/ph14100966. Pharmaceuticals (Basel). 2021. PMID: 34681191 Free PMC article.
-
Factor XI in Carriers of Antiphospholipid Antibodies: Elevated Levels Associated with Symptomatic Thrombotic Cases, While Low Levels Linked to Asymptomatic Cases.Int J Mol Sci. 2023 Nov 13;24(22):16270. doi: 10.3390/ijms242216270. Int J Mol Sci. 2023. PMID: 38003459 Free PMC article.
-
High-Throughput Docking and Molecular Dynamics Simulations towards the Identification of Potential Inhibitors against Human Coagulation Factor XIIa.Comput Math Methods Med. 2020 May 22;2020:2852051. doi: 10.1155/2020/2852051. eCollection 2020. Comput Math Methods Med. 2020. PMID: 32549905 Free PMC article.
-
Allosteric inhibition of factor XIa. Sulfated non-saccharide glycosaminoglycan mimetics as promising anticoagulants.Thromb Res. 2015 Aug;136(2):379-87. doi: 10.1016/j.thromres.2015.04.017. Epub 2015 Apr 22. Thromb Res. 2015. PMID: 25935648 Free PMC article.
References
-
- Furie B, Furie BC. Molecular basis of blood coagulation. In: Hoffman H, Benz EJ, Shattil SJ, et al., editors. Hematology, Basic Principles and Practice. 5th ed. Philadelphia, PA: Churchill Livingstone-Elsevier; 2009. pp. 1819–1836.
-
- Seligsohn U. Factor XI in haemostasis and thrombosis: past, present and future. Thromb Haemost. 2007;98(1):84–89. - PubMed
-
- Bouma BN, Griffin JH. Human blood coagulation factor XI: purification, properties, and mechanism of activation by activated factor XII. J Biol Chem. 1977;252(18):6432–6437. - PubMed
-
- Fujikawa K, Chung DW, Hendrickson LE, Davie EW. Amino acid sequence of human factor XI, a blood coagulation factor with four tandem repeats that are highly homologous with plasma prekallikrein. Biochemistry. 1986;25(9):2417–2424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

