Expansion of the eukaryotic proteome by alternative splicing
- PMID: 20110989
- PMCID: PMC3443858
- DOI: 10.1038/nature08909
Expansion of the eukaryotic proteome by alternative splicing
Abstract
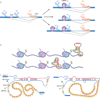
The collection of components required to carry out the intricate processes involved in generating and maintaining a living, breathing and, sometimes, thinking organism is staggeringly complex. Where do all of the parts come from? Early estimates stated that about 100,000 genes would be required to make up a mammal; however, the actual number is less than one-quarter of that, barely four times the number of genes in budding yeast. It is now clear that the 'missing' information is in large part provided by alternative splicing, the process by which multiple different functional messenger RNAs, and therefore proteins, can be synthesized from a single gene.
Figures


References
-
- Alt FW, et al. Synthesis of secreted and membrane-bound immunoglobulin μ heavy chains is directed by mRNAs that differ at their 3’ ends. Cell. 1980;20:293–301. - PubMed
-
- Early P, et al. Two mRNAs can be produced from a single immunoglobulin μ gene by alternative RNA processing pathways. Cell. 1980;20:313–319. - PubMed
-
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 2008;40:1413–1415. - PubMed
-
-
Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. References and provide detailed views of the human transcriptome as determined by using deep-sequencing data. The authors conclude that the pre-mRNAs from all multi- exon genes are alternatively spliced.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

