Chromatin remodelling during development
- PMID: 20110991
- PMCID: PMC3060774
- DOI: 10.1038/nature08911
Chromatin remodelling during development
Abstract
New methods for the genome-wide analysis of chromatin are providing insight into its roles in development and their underlying mechanisms. Current studies indicate that chromatin is dynamic, with its structure and its histone modifications undergoing global changes during transitions in development and in response to extracellular cues. In addition to DNA methylation and histone modification, ATP-dependent enzymes that remodel chromatin are important controllers of chromatin structure and assembly, and are major contributors to the dynamic nature of chromatin. Evidence is emerging that these chromatin-remodelling enzymes have instructive and programmatic roles during development. Particularly intriguing are the findings that specialized assemblies of ATP-dependent remodellers are essential for establishing and maintaining pluripotent and multipotent states in cells.
Figures
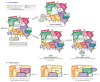
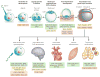

References
-
- Kornberg RD. Chromatin structure: a repeating unit of histones and DNA. Science. 1974;184:868–871. - PubMed
-
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nature Rev Genet. 2008;9:465–476. - PubMed
-
- Wang Y, et al. Linking covalent histone modifications to epigenetics: the rigidity and plasticity of the marks. Cold Spring Harb Symp Quant Biol. 2004;69:161–169. - PubMed
-
- Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

