RNA microarray analysis in prenatal mouse cochlea reveals novel IGF-I target genes: implication of MEF2 and FOXM1 transcription factors
- PMID: 20111592
- PMCID: PMC2810322
- DOI: 10.1371/journal.pone.0008699
RNA microarray analysis in prenatal mouse cochlea reveals novel IGF-I target genes: implication of MEF2 and FOXM1 transcription factors
Abstract
Background: Insulin-like growth factor-I (IGF-I) provides pivotal cell survival and differentiation signals during inner ear development throughout evolution. Homozygous mutations of human IGF1 cause syndromic sensorineural deafness, decreased intrauterine and postnatal growth rates, and mental retardation. In the mouse, deficits in IGF-I result in profound hearing loss associated with reduced survival, differentiation and maturation of auditory neurons. Nevertheless, little is known about the molecular basis of IGF-I activity in hearing and deafness.
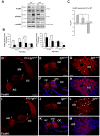
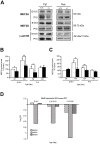
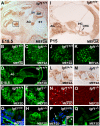
Methodology/principal findings: A combination of quantitative RT-PCR, subcellular fractionation and Western blotting, along with in situ hybridization studies show IGF-I and its high affinity receptor to be strongly expressed in the embryonic and postnatal mouse cochlea. The expression of both proteins decreases after birth and in the cochlea of E18.5 embryonic Igf1(-/-) null mice, the balance of the main IGF related signalling pathways is altered, with lower activation of Akt and ERK1/2 and stronger activation of p38 kinase. By comparing the Igf1(-/-) and Igf1(+/+) transcriptomes in E18.5 mouse cochleae using RNA microchips and validating their results, we demonstrate the up-regulation of the FoxM1 transcription factor and the misexpression of the neural progenitor transcription factors Six6 and Mash1 associated with the loss of IGF-I. Parallel, in silico promoter analysis of the genes modulated in conjunction with the loss of IGF-I revealed the possible involvement of MEF2 in cochlear development. E18.5 Igf1(+/+) mouse auditory ganglion neurons showed intense MEF2A and MEF2D nuclear staining and MEF2A was also evident in the organ of Corti. At P15, MEF2A and MEF2D expression were shown in neurons and sensory cells. In the absence of IGF-I, nuclear levels of MEF2 were diminished, indicating less transcriptional MEF2 activity. By contrast, there was an increase in the nuclear accumulation of FoxM1 and a corresponding decrease in the nuclear cyclin-dependent kinase inhibitor p27(Kip1).
Conclusions/significance: We have defined the spatiotemporal expression of elements involved in IGF signalling during inner ear development and reveal novel regulatory mechanisms that are modulated by IGF-I in promoting sensory cell and neural survival and differentiation. These data will help us to understand the molecular bases of human sensorineural deafness associated to deficits in IGF-I.
Conflict of interest statement
Figures





References
-
- Piper MD, Selman C, McElwee JJ, Partridge L. Separating cause from effect: how does insulin/IGF signalling control lifespan in worms, flies and mice? J Intern Med. 2008;263:179–191. - PubMed
-
- Russo VC, Gluckman PD, Feldman EL, Werther GA. The insulin-like growth factor system and its pleiotropic functions in brain. Endocr Rev. 2005;26:916–943. - PubMed
-
- Werner H, Weinstein D, Bentov I. Similarities and differences between insulin and IGF-I: structures, receptors, and signalling pathways. Arch Physiol Biochem. 2008;114:17–22. - PubMed
-
- LeRoith D. Insulin-like growth factors and the brain. Endocrinology. 2008;149:5951. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

