Mutational analysis of highly conserved residues in the phage phiC31 integrase reveals key amino acids necessary for the DNA recombination
- PMID: 20111606
- PMCID: PMC2810336
- DOI: 10.1371/journal.pone.0008863
Mutational analysis of highly conserved residues in the phage phiC31 integrase reveals key amino acids necessary for the DNA recombination
Abstract
Background: Amino acid sequence alignment of phage phiC31 integrase with the serine recombinases family revealed highly conserved regions outside the catalytic domain. Until now, no system mutational or biochemical studies have been carried out to assess the roles of these conserved residues in the recombination of phiC31 integrase.
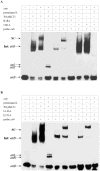
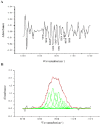
Methodology/principal findings: To determine the functional roles of these conserved residues, a series of conserved residues were targeted by site-directed mutagenesis. Out of the 17 mutants, 11 mutants showed impaired or no recombination ability, as analyzed by recombination assay both in vivo and in vitro. Results of DNA binding activity assays showed that mutants (R18A, I141A, L143A,E153A, I432A and V571A) exhibited a great decrease in DNA binding affinity, and mutants (G182A/F183A, C374A, C376A/G377A, Y393A and V566A) had completely lost their ability to bind to the specific target DNA attB as compared with wild-type protein. Further analysis of mutants (R18A, I141A, L143A and E153A) synapse and cleavage showed that these mutants were blocked in recombination at the stage of strand cleavage.
Conclusions/significance: This data reveals that some of the highly conserved residues both in the N-terminus and C-terminus region of phiC31 integrase, play vital roles in the substrate binding and cleavage. The cysteine-rich motif and the C-tail val-rich region of phiC31 integrase may represent the major DNA binding domains of phiC31 integrase.
Conflict of interest statement
Figures





Similar articles
-
Critical amino acid residues within the φC31 integrase DNA-binding domain affect recombination activities in mammalian cells.Hum Gene Ther. 2010 Sep;21(9):1104-18. doi: 10.1089/hum.2010.034. Hum Gene Ther. 2010. PMID: 20415519
-
Synapsis and DNA cleavage in phiC31 integrase-mediated site-specific recombination.Nucleic Acids Res. 2004 May 11;32(8):2607-17. doi: 10.1093/nar/gkh538. Print 2004. Nucleic Acids Res. 2004. PMID: 15141031 Free PMC article.
-
Preliminary study on the DNA-binding properties of phage ΦC31 integrase.Gene. 2011 Sep 15;484(1-2):47-51. doi: 10.1016/j.gene.2011.05.020. Epub 2011 Jun 12. Gene. 2011. PMID: 21679753
-
Site-specific recombination by phiC31 integrase and other large serine recombinases.Biochem Soc Trans. 2010 Apr;38(2):388-94. doi: 10.1042/BST0380388. Biochem Soc Trans. 2010. PMID: 20298189 Review.
-
Phage-encoded Serine Integrases and Other Large Serine Recombinases.Microbiol Spectr. 2015 Aug;3(4). doi: 10.1128/microbiolspec.MDNA3-0059-2014. Microbiol Spectr. 2015. PMID: 26350324 Review.
Cited by
-
Streptomyces temperate bacteriophage integration systems for stable genetic engineering of actinomycetes (and other organisms).J Ind Microbiol Biotechnol. 2012 May;39(5):661-72. doi: 10.1007/s10295-011-1069-6. Epub 2011 Dec 13. J Ind Microbiol Biotechnol. 2012. PMID: 22160317 Review.
-
Pseudo attP sites in favor of transgene integration and expression in cultured porcine cells identified by Streptomyces phage phiC31 integrase.BMC Mol Biol. 2013 Sep 8;14:20. doi: 10.1186/1471-2199-14-20. BMC Mol Biol. 2013. PMID: 24010979 Free PMC article.
-
Nanoparticles for retinal gene therapy.Prog Retin Eye Res. 2010 Sep;29(5):376-97. doi: 10.1016/j.preteyeres.2010.04.004. Epub 2010 May 7. Prog Retin Eye Res. 2010. PMID: 20452457 Free PMC article. Review.
-
A bipartite thermodynamic-kinetic contribution by an activating mutation to RDF-independent excision by a phage serine integrase.Nucleic Acids Res. 2020 Jul 9;48(12):6413-6430. doi: 10.1093/nar/gkaa401. Nucleic Acids Res. 2020. PMID: 32479633 Free PMC article.
-
Single-molecule analysis of ϕC31 integrase-mediated site-specific recombination by tethered particle motion.Nucleic Acids Res. 2016 Dec 15;44(22):10804-10823. doi: 10.1093/nar/gkw861. Epub 2016 Oct 5. Nucleic Acids Res. 2016. PMID: 27986956 Free PMC article.
References
-
- Kuhstoss S, Rao RN. Analysis of the integration function of the streptomycete bacteriophage phiC31. J Mol Biol. 1991;222:897–908. - PubMed
-
- Thomason LC, Calendar R, Ow DW. Gene insertion and replacement in Schizosaccharomyces pombe mediated by the Streptomyces bacteriophage phiC31 site-specific recombination system. Mol Genet Genomics. 2001;265:1031–1038. - PubMed
-
- Grainge I, Pathania S, Vologodskii A, Harshey RM, Jayaram M. Symmetric DNA sites are functionally asymmetric within Flp and Cre site-specific DNA recombination synapses. J Mol Biol. 2002;320:515–527. - PubMed
-
- Groth AC, Calos MP. Phage integrases: biology and applications. J Mol Biol. 2004;335:667–678. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

