The zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei
- PMID: 20111718
- PMCID: PMC2811742
- DOI: 10.1371/journal.pone.0008913
The zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei
Abstract
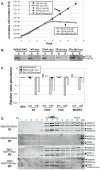
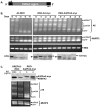
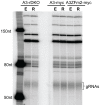
Most mitochondrial mRNAs in trypanosomes undergo uridine insertion/deletion editing that is catalyzed by approximately 20S editosomes. The editosome component KREPA3 is essential for editosome structural integrity and its two zinc finger (ZF) motifs are essential for editing in vivo but not in vitro. KREPA3 function was further explored by examining the consequence of mutation of its N- and C-terminal ZFs (ZF1 and ZF2, respectively). Exclusively expressed myc-tagged KREPA3 with ZF2 mutation resulted in lower KREPA3 abundance and a relative increase in KREPA2 and KREL1 proteins. Detailed analysis of edited RNA products revealed the accumulation of partially edited mRNAs with less insertion editing compared to the partially edited mRNAs found in the cells with wild type KREPA3 expression. Mutation of ZF1 in TAP-tagged KREPA3 also resulted in accumulation of partially edited mRNAs that were shorter and only edited in the 3'-terminal editing region. Mutation of both ZFs essentially eliminated partially edited mRNA. The mutations did not affect gRNA abundance. These data indicate that both ZFs are essential for the progression of editing and perhaps its accuracy, which suggests that KREPA3 plays roles in the editing process via its ZFs interaction with editosome proteins and/or RNA substrates.
Conflict of interest statement
Figures



Similar articles
-
Differential Editosome Protein Function between Life Cycle Stages of Trypanosoma brucei.J Biol Chem. 2015 Oct 9;290(41):24914-31. doi: 10.1074/jbc.M115.669432. Epub 2015 Aug 24. J Biol Chem. 2015. PMID: 26304125 Free PMC article.
-
The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei.Mol Cell Biol. 2008 Nov;28(22):6939-53. doi: 10.1128/MCB.01115-08. Epub 2008 Sep 15. Mol Cell Biol. 2008. PMID: 18794366 Free PMC article.
-
Mutational analysis of Trypanosoma brucei editosome proteins KREPB4 and KREPB5 reveals domains critical for function.RNA. 2012 Oct;18(10):1897-909. doi: 10.1261/rna.035048.112. Epub 2012 Aug 23. RNA. 2012. PMID: 22919050 Free PMC article.
-
Dynamic RNA holo-editosomes with subcomplex variants: Insights into the control of trypanosome editing.Wiley Interdiscip Rev RNA. 2018 Nov;9(6):e1502. doi: 10.1002/wrna.1502. Epub 2018 Aug 12. Wiley Interdiscip Rev RNA. 2018. PMID: 30101566 Free PMC article. Review.
-
Structure and intrinsic disorder of the proteins of the Trypanosoma brucei editosome.FEBS Lett. 2015 Sep 14;589(19 Pt A):2603-10. doi: 10.1016/j.febslet.2015.07.026. Epub 2015 Jul 29. FEBS Lett. 2015. PMID: 26226426 Review.
Cited by
-
Inhibitors of RNA editing as potential chemotherapeutics against trypanosomatid pathogens.Int J Parasitol Drugs Drug Resist. 2011 Nov 13;2:36-46. doi: 10.1016/j.ijpddr.2011.10.003. eCollection 2012 Dec. Int J Parasitol Drugs Drug Resist. 2011. PMID: 24533263 Free PMC article. Review.
-
Differential Editosome Protein Function between Life Cycle Stages of Trypanosoma brucei.J Biol Chem. 2015 Oct 9;290(41):24914-31. doi: 10.1074/jbc.M115.669432. Epub 2015 Aug 24. J Biol Chem. 2015. PMID: 26304125 Free PMC article.
-
High-throughput sequencing of partially edited trypanosome mRNAs reveals barriers to editing progression and evidence for alternative editing.RNA. 2016 May;22(5):677-95. doi: 10.1261/rna.055160.115. Epub 2016 Feb 23. RNA. 2016. PMID: 26908922 Free PMC article.
-
Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer.Wiley Interdiscip Rev RNA. 2011 Sep-Oct;2(5):669-85. doi: 10.1002/wrna.82. Epub 2011 Mar 23. Wiley Interdiscip Rev RNA. 2011. PMID: 21823228 Free PMC article. Review.
-
Explorations of linked editosome domains leading to the discovery of motifs defining conserved pockets in editosome OB-folds.J Struct Biol. 2012 Nov;180(2):362-73. doi: 10.1016/j.jsb.2012.07.012. Epub 2012 Aug 10. J Struct Biol. 2012. PMID: 22902563 Free PMC article.
References
-
- Benne R. RNA editing in trypanosomes. Eur J Biochem. 1994;221:9–23. - PubMed
-
- Lukes J, Hashimi H, Zikova A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr Genet. 2005;48:277–299. - PubMed
-
- Bhat GJ, Koslowsky DJ, Feagin JE, Smiley BL, Stuart K. An extensively edited mitochondrial transcript in kinetoplastids encodes a protein homologous to ATPase subunit 6. Cell. 1990;61:885–894. - PubMed
-
- Feagin JE, Abraham JM, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

