Molecular analysis of vector genome structures after liver transduction by conventional and self-complementary adeno-associated viral serotype vectors in murine and nonhuman primate models
- PMID: 20113166
- PMCID: PMC2938357
- DOI: 10.1089/hum.2009.214
Molecular analysis of vector genome structures after liver transduction by conventional and self-complementary adeno-associated viral serotype vectors in murine and nonhuman primate models
Abstract
Vectors based on several new adeno-associated viral (AAV) serotypes demonstrated strong hepatocyte tropism and transduction efficiency in both small- and large-animal models for liver-directed gene transfer. Efficiency of liver transduction by AAV vectors can be further improved in both murine and nonhuman primate (NHP) animals when the vector genomes are packaged in a self-complementary (sc) format. In an attempt to understand potential molecular mechanism(s) responsible for enhanced transduction efficiency of the sc vector in liver, we performed extensive molecular studies of genome structures of conventional single-stranded (ss) and sc AAV vectors from liver after AAV gene transfer in both mice and NHPs. These included treatment with exonucleases with specific substrate preferences, single-cutter restriction enzyme digestion and polarity-specific hybridization-based vector genome mapping, and bacteriophage phi29 DNA polymerase-mediated and double-stranded circular template-specific rescue of persisted circular genomes. In mouse liver, vector genomes of both genome formats seemed to persist primarily as episomal circular forms, but sc vectors converted into circular forms more rapidly and efficiently. However, the overall differences in vector genome abundance and structure in the liver between ss and sc vectors could not account for the remarkable differences in transduction. Molecular structures of persistent genomes of both ss and sc vectors were significantly more heterogeneous in macaque liver, with noticeable structural rearrangements that warrant further characterizations.
Figures

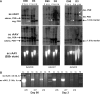
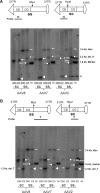



Similar articles
-
Optimized adeno-associated virus (AAV)-protein phosphatase-5 helper viruses for efficient liver transduction by single-stranded AAV vectors: therapeutic expression of factor IX at reduced vector doses.Hum Gene Ther. 2010 Mar;21(3):271-83. doi: 10.1089/hum.2009.100. Hum Gene Ther. 2010. PMID: 19788390 Free PMC article.
-
Self-complementary adeno-associated virus vectors containing a novel liver-specific human factor IX expression cassette enable highly efficient transduction of murine and nonhuman primate liver.Blood. 2006 Apr 1;107(7):2653-61. doi: 10.1182/blood-2005-10-4035. Epub 2005 Dec 1. Blood. 2006. PMID: 16322469 Free PMC article.
-
Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors.J Virol. 2004 Mar;78(6):3110-22. doi: 10.1128/jvi.78.6.3110-3122.2004. J Virol. 2004. PMID: 14990730 Free PMC article.
-
New recombinant serotypes of AAV vectors.Curr Gene Ther. 2005 Jun;5(3):285-97. doi: 10.2174/1566523054065057. Curr Gene Ther. 2005. PMID: 15975006 Review.
-
Adeno-associated viral vectors for correction of inborn errors of metabolism: progressing towards clinical application.Curr Pharm Des. 2011;17(24):2500-15. doi: 10.2174/138161211797247569. Curr Pharm Des. 2011. PMID: 21774772 Review.
Cited by
-
Adeno-associated virus type 2 (AAV2) uncoating is a stepwise process and is linked to structural reorganization of the nucleolus.PLoS Pathog. 2022 Jul 11;18(7):e1010187. doi: 10.1371/journal.ppat.1010187. eCollection 2022 Jul. PLoS Pathog. 2022. PMID: 35816507 Free PMC article.
-
Integration frequency and intermolecular recombination of rAAV vectors in non-human primate skeletal muscle and liver.Mol Ther. 2012 Jun;20(6):1177-86. doi: 10.1038/mt.2012.47. Epub 2012 Mar 27. Mol Ther. 2012. PMID: 22453768 Free PMC article.
-
Interferon-γ inducible factor 16 (IFI16) restricts adeno-associated virus type 2 (AAV2) transduction in an immune-modulatory independent way.J Virol. 2024 Jul 23;98(7):e0011024. doi: 10.1128/jvi.00110-24. Epub 2024 Jun 5. J Virol. 2024. PMID: 38837381 Free PMC article.
-
Recombinant Adeno-Associated Virus Vector Genomes Take the Form of Long-Lived, Transcriptionally Competent Episomes in Human Muscle.Hum Gene Ther. 2016 Jan;27(1):32-42. doi: 10.1089/hum.2015.136. Hum Gene Ther. 2016. PMID: 26650966 Free PMC article.
-
Herpes Simplex Virus 1 Coinfection Modifies Adeno-associated Virus Genome End Recombination.J Virol. 2021 Jun 10;95(13):e0048621. doi: 10.1128/JVI.00486-21. Epub 2021 Jun 10. J Virol. 2021. PMID: 33853961 Free PMC article.
References
-
- Alexander I.E. Cunningham S.C. Logan G.J. Christodoulou J. Potential of AAV vectors in the treatment of metabolic disease. Gene Ther. 2008;15:831–839. - PubMed
-
- Davidoff A.M. Gray J.T. Ng C.Y. Zhang Y. Zhou J. Spence Y. Bakar Y. Nathwani A.C. Comparison of the ability of adeno-associated viral vectors pseudotyped with serotype 2, 5, and 8 capsid proteins to mediate efficient transduction of the liver in murine and nonhuman primate models. Mol. Ther. 2005;11:875–888. - PubMed
-
- DeLong E.F. Preston C.M. Mincer T. Rich V. Hallam S.J. Frigaard N.U. Martinez A. Sullivan M.B. Edwards R. Brito B.R. Chisholm S.W. Karl D.M. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

