Orthophosphate binding at the dimer interface of Corynebacterium callunae starch phosphorylase: mutational analysis of its role for activity and stability of the enzyme
- PMID: 20113461
- PMCID: PMC2837607
- DOI: 10.1186/1471-2091-11-8
Orthophosphate binding at the dimer interface of Corynebacterium callunae starch phosphorylase: mutational analysis of its role for activity and stability of the enzyme
Abstract
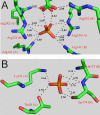
Background: Orthophosphate recognition at allosteric binding sites is a key feature for the regulation of enzyme activity in mammalian glycogen phosphorylases. Protein residues co-ordinating orthophosphate in three binding sites distributed across the dimer interface of a non-regulated bacterial starch phosphorylase (from Corynebacterium callunae) were individually replaced by Ala to interrogate their unknown function for activity and stability of this enzyme.
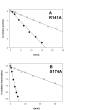
Results: While the mutations affected neither content of pyridoxal 5'-phosphate cofactor nor specific activity in phosphorylase preparations as isolated, they disrupted (Thr28-->Ala, Arg141-->Ala) or decreased (Lys31-->Ala, Ser174-->Ala) the unusually strong protective effect of orthophosphate (10 or 100 mM) against inactivation at 45 degrees C and subunit dissociation enforced by imidazole, as compared to wild-type enzyme. Loss of stability in the mutated phosphorylases appeared to be largely due to weakened affinity for orthophosphate binding. Binding of sulphate mimicking the crystallographically observed "non-covalent phosphorylation" of the phosphorylase at the dimer interface did not have an allosteric effect on the enzyme activity.
Conclusions: The phosphate sites at the subunit-subunit interface of C. callunae starch phosphorylase appear to be cooperatively functional in conferring extra kinetic stability to the native dimer structure of the active enzyme. The molecular strategy exploited for quaternary structure stabilization is to our knowledge novel among dimeric proteins. It can be distinguished clearly from the co-solute effect of orthophosphate on protein thermostability resulting from (relatively weak) interactions of the ligand with protein surface residues.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

