Proteomic comparison of the cytosolic proteins of three Bifidobacterium longum human isolates and B. longum NCC2705
- PMID: 20113481
- PMCID: PMC2824696
- DOI: 10.1186/1471-2180-10-29
Proteomic comparison of the cytosolic proteins of three Bifidobacterium longum human isolates and B. longum NCC2705
Abstract
Background: Bifidobacteria are natural inhabitants of the human gastrointestinal tract. In full-term newborns, these bacteria are acquired from the mother during delivery and rapidly become the predominant organisms in the intestinal microbiota. Bifidobacteria contribute to the establishment of healthy intestinal ecology and can confer health benefits to their host. Consequently, there is growing interest in bifidobacteria, and various strains are currently used as probiotic components in functional food products. However, the probiotic effects have been reported to be strain-specific. There is thus a need to better understand the determinants of the observed benefits provided by these probiotics. Our objective was to compare three human B. longum isolates with the sequenced model strain B. longum NCC2705 at the chromosome and proteome levels.
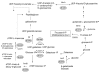
Results: Pulsed field electrophoresis genotyping revealed genetic heterogeneity with low intraspecies strain relatedness among the four strains tested. Using two-dimensional gel electrophoresis, we analyzed qualitative differences in the cytosolic protein patterns. There were 45 spots that were present in some strains and absent in others. Spots were excised from the gels and subjected to peptide mass fingerprint analysis for identification. The 45 spots represented 37 proteins, most of which were involved in carbohydrate metabolism and cell wall or cell membrane synthesis. Notably, the protein patterns were correlated with differences in cell membrane properties like surface hydrophobicity and cell agglutination.
Conclusion: These results showed that proteomic analysis can be valuable for investigating differences in bifidobacterial species and may provide a better understanding of the diversity of bifidobacteria and their potential use as probiotics.
Figures

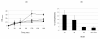
References
-
- Bezkorovainy A. Probiotics: determinants of survival and growth in the gut. Am J Clin Nutr. 2001;73:399S–405S. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

