Transcriptome analysis of the Cryptocaryon irritans tomont stage identifies potential genes for the detection and control of cryptocaryonosis
- PMID: 20113487
- PMCID: PMC2828411
- DOI: 10.1186/1471-2164-11-76
Transcriptome analysis of the Cryptocaryon irritans tomont stage identifies potential genes for the detection and control of cryptocaryonosis
Abstract
Background: Cryptocaryon irritans is a parasitic ciliate that causes cryptocaryonosis (white spot disease) in marine fish. Diagnosis of cryptocaryonosis often depends on the appearance of white spots on the surface of the fish, which are usually visible only during later stages of the disease. Identifying suitable biomarkers of this parasite would aid the development of diagnostic tools and control strategies for C. irritans. The C. irritans genome is virtually unexplored; therefore, we generated and analyzed expressed sequence tags (ESTs) of the parasite to identify genes that encode for surface proteins, excretory/secretory proteins and repeat-containing proteins.
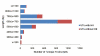
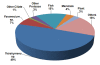
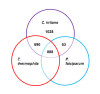
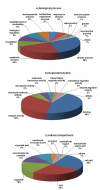
Results: ESTs were generated from a cDNA library of C. irritans tomonts isolated from infected Asian sea bass, Lates calcarifer. Clustering of the 5356 ESTs produced 2659 unique transcripts (UTs) containing 1989 singletons and 670 consensi. BLAST analysis showed that 74% of the UTs had significant similarity (E-value < 10-5) to sequences that are currently available in the GenBank database, with more than 15% of the significant hits showing unknown function. Forty percent of the UTs had significant similarity to ciliates from the genera Tetrahymena and Paramecium. Comparative gene family analysis with related taxa showed that many protein families are conserved among the protozoans. Based on gene ontology annotation, functional groups were successfully assigned to 790 UTs. Genes encoding excretory/secretory proteins and membrane and membrane-associated proteins were identified because these proteins often function as antigens and are good antibody targets. A total of 481 UTs were classified as encoding membrane proteins, 54 were classified as encoding for membrane-bound proteins, and 155 were found to contain excretory/secretory protein-coding sequences. Amino acid repeat-containing proteins and GPI-anchored proteins were also identified as potential candidates for the development of diagnostic and control strategies for C. irritans.
Conclusions: We successfully discovered and examined a large portion of the previously unexplored C. irritans transcriptome and identified potential genes for the development and validation of diagnostic and control strategies for cryptocaryonosis.
Figures

References
-
- Colorni A, Burgess P. Cryptocaryon irritans Brown 1951, the cause of 'white spot disease' in marine fish: an update. Aquar Sci Conserv. 1997;1(4):217–238. doi: 10.1023/A:1018360323287. - DOI
-
- Diggles BK, Lester RJG. Infections of Cryptocaryon irritans on wild fish from southeast Queensland, Australia. Dis Aquat Organ. 1996;25(3):159–167. doi: 10.3354/dao025159. - DOI
-
- Diamant A, Issar G, Colorni A, Paperna I. A pathogenic Cryptocaryon-like ciliate from the Mediterranean Sea. Bull Eur Assoc Fish Pathol. 1991;11(3):122–124.
-
- Colorni A. Aspects of the biology of Cryptocaryon irritans, and hyposalinity as a control measure in gilt-head sea bream Sparus aurata. Dis Aquat Organ. 1985;1:19–22. doi: 10.3354/dao001019. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

