A temporal precedence based clustering method for gene expression microarray data
- PMID: 20113513
- PMCID: PMC2841598
- DOI: 10.1186/1471-2105-11-68
A temporal precedence based clustering method for gene expression microarray data
Abstract
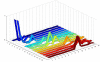
Background: Time-course microarray experiments can produce useful data which can help in understanding the underlying dynamics of the system. Clustering is an important stage in microarray data analysis where the data is grouped together according to certain characteristics. The majority of clustering techniques are based on distance or visual similarity measures which may not be suitable for clustering of temporal microarray data where the sequential nature of time is important. We present a Granger causality based technique to cluster temporal microarray gene expression data, which measures the interdependence between two time-series by statistically testing if one time-series can be used for forecasting the other time-series or not.
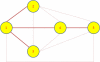
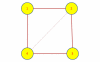
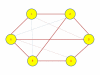
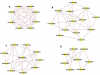
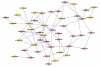
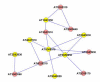
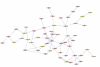
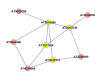
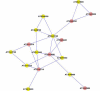
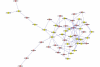
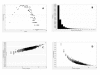
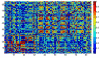
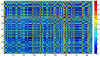
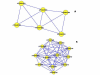
Results: A gene-association matrix is constructed by testing temporal relationships between pairs of genes using the Granger causality test. The association matrix is further analyzed using a graph-theoretic technique to detect highly connected components representing interesting biological modules. We test our approach on synthesized datasets and real biological datasets obtained for Arabidopsis thaliana. We show the effectiveness of our approach by analyzing the results using the existing biological literature. We also report interesting structural properties of the association network commonly desired in any biological system.
Conclusions: Our experiments on synthesized and real microarray datasets show that our approach produces encouraging results. The method is simple in implementation and is statistically traceable at each step. The method can produce sets of functionally related genes which can be further used for reverse-engineering of gene circuits.
Figures



Similar articles
-
Listen to genes: dealing with microarray data in the frequency domain.PLoS One. 2009;4(4):e5098. doi: 10.1371/journal.pone.0005098. Epub 2009 Apr 6. PLoS One. 2009. PMID: 22745650 Free PMC article.
-
Proximity measures for clustering gene expression microarray data: a validation methodology and a comparative analysis.IEEE/ACM Trans Comput Biol Bioinform. 2013 Jul-Aug;10(4):845-57. doi: 10.1109/TCBB.2013.9. IEEE/ACM Trans Comput Biol Bioinform. 2013. PMID: 24334380
-
Multiple gene expression profile alignment for microarray time-series data clustering.Bioinformatics. 2010 Sep 15;26(18):2281-8. doi: 10.1093/bioinformatics/btq422. Epub 2010 Jul 16. Bioinformatics. 2010. PMID: 20639411
-
Manipulating large-scale Arabidopsis microarray expression data: identifying dominant expression patterns and biological process enrichment.Methods Mol Biol. 2009;553:57-77. doi: 10.1007/978-1-60327-563-7_4. Methods Mol Biol. 2009. PMID: 19588101 Free PMC article. Review.
-
Computational approaches to analysis of DNA microarray data.Yearb Med Inform. 2006:91-103. Yearb Med Inform. 2006. PMID: 17051302 Review.
Cited by
-
Functional clustering of time series gene expression data by Granger causality.BMC Syst Biol. 2012 Oct 30;6:137. doi: 10.1186/1752-0509-6-137. BMC Syst Biol. 2012. PMID: 23107425 Free PMC article.
-
Dysregulated cellular redox status during hyperammonemia causes mitochondrial dysfunction and senescence by inhibiting sirtuin-mediated deacetylation.Aging Cell. 2023 Jul;22(7):e13852. doi: 10.1111/acel.13852. Epub 2023 Apr 26. Aging Cell. 2023. PMID: 37101412 Free PMC article.
-
Time-series clustering of gene expression in irradiated and bystander fibroblasts: an application of FBPA clustering.BMC Genomics. 2011 Jan 4;12:2. doi: 10.1186/1471-2164-12-2. BMC Genomics. 2011. PMID: 21205307 Free PMC article.
References
-
- Kim BR, Littell RC, Wu RL. Clustering the periodic pattern of gene expression using Fourier series approximations. Curr Genomics. 2006;7:197–203. doi: 10.2174/138920206777780229. - DOI
-
- Speed T. Statistical Analysis of Gene Expression Microarray Data. Chapman and Hall/CRC; 2003.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

