Differential roles of cyclin D1 and D3 in pancreatic ductal adenocarcinoma
- PMID: 20113529
- PMCID: PMC2824633
- DOI: 10.1186/1476-4598-9-24
Differential roles of cyclin D1 and D3 in pancreatic ductal adenocarcinoma
Abstract
Background: The cyclin D1 (CCND1) and cyclin D3 (CCND3) are frequently co-overexpressed in pancreatic ductal adenocarcinoma (PDAC). Here we examine their differential roles in PDAC.
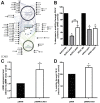
Results: CCND1 and CCND3 expression were selectively suppressed by shRNA in PDAC cell lines with expression levels of equal CCND1 and CCND3 (BxPC3), enhanced CCND1 (HPAC) or enhanced CCND3 (PANC1). Suppression of cell proliferation was greater with CCND3 than CCND1 downregulation. CCND3 suppression led to a reduced level of phosphorylated retinoblastoma protein (Ser795p-Rb/p110) and resulted in decreased levels of cyclin A mRNA and protein. A global gene expression analysis identified deregulated genes in D1- or D3-cyclin siRNA-treated PANC1 cells. The downregulated gene targets in CCND3 suppressed cells were significantly enriched in cell cycle associated processes (p < 0.005). In contrast, focal adhesion/actin cytoskeleton, MAPK and NF B signaling appeared to characterize the target genes and their interacting proteins in CCND1 suppressed PANC1 cells.
Conclusions: Our results suggest that CCND3 is the primary driver of the cell cycle, in cooperation with CCND1 that integrates extracellular mitogenic signaling. We also present evidence that CCND1 plays a role in tumor cell migration. The results provide novel insights for common and differential targets of CCND1 and CCND3 overexpression during pancreatic duct cell carcinogenesis.
Figures




Similar articles
-
Overexpression of G1-S cyclins and cyclin-dependent kinases during multistage human pancreatic duct cell carcinogenesis.Clin Cancer Res. 2004 Oct 1;10(19):6598-605. doi: 10.1158/1078-0432.CCR-04-0524. Clin Cancer Res. 2004. PMID: 15475449
-
Downregulated expression of IL‑28RA is involved in the pathogenesis of pancreatic ductal adenocarcinoma.Int J Oncol. 2021 Aug;59(2):55. doi: 10.3892/ijo.2021.5235. Epub 2021 Jul 1. Int J Oncol. 2021. PMID: 34195850
-
Syk tyrosine kinase acts as a pancreatic adenocarcinoma tumor suppressor by regulating cellular growth and invasion.Am J Pathol. 2009 Dec;175(6):2625-36. doi: 10.2353/ajpath.2009.090543. Epub 2009 Nov 5. Am J Pathol. 2009. PMID: 19893036 Free PMC article.
-
Amplification and expression of cyclin D genes (CCND1, CCND2 and CCND3) in human malignant gliomas.Brain Pathol. 1999 Jul;9(3):435-42; discussion 432-3. doi: 10.1111/j.1750-3639.1999.tb00532.x. Brain Pathol. 1999. PMID: 10416984 Free PMC article.
-
An update on the implications of cyclin D1 in oral carcinogenesis.Oral Dis. 2017 Oct;23(7):897-912. doi: 10.1111/odi.12620. Epub 2017 Mar 31. Oral Dis. 2017. PMID: 27973759 Review.
Cited by
-
The Essential Role of Rac1 Glucosylation in Clostridioides difficile Toxin B-Induced Arrest of G1-S Transition.Front Microbiol. 2022 Mar 7;13:846215. doi: 10.3389/fmicb.2022.846215. eCollection 2022. Front Microbiol. 2022. PMID: 35321078 Free PMC article.
-
Modified gateway system for double shRNA expression and Cre/lox based gene expression.BMC Biotechnol. 2011 Mar 22;11:24. doi: 10.1186/1472-6750-11-24. BMC Biotechnol. 2011. PMID: 21418658 Free PMC article.
-
Translational repression of cyclin D3 by a stable G-quadruplex in its 5' UTR: implications for cell cycle regulation.RNA Biol. 2012 Aug;9(8):1099-109. doi: 10.4161/rna.21210. Epub 2012 Aug 1. RNA Biol. 2012. PMID: 22858673 Free PMC article.
-
MicroRNA-195 plays a tumor-suppressor role in human glioblastoma cells by targeting signaling pathways involved in cellular proliferation and invasion.Neuro Oncol. 2012 Mar;14(3):278-87. doi: 10.1093/neuonc/nor216. Epub 2012 Jan 4. Neuro Oncol. 2012. PMID: 22217655 Free PMC article.
-
BCL9L expression in pancreatic neoplasia with a focus on SPN: a possible explanation for the enigma of the benign neoplasia.BMC Cancer. 2016 Aug 18;16:648. doi: 10.1186/s12885-016-2707-1. BMC Cancer. 2016. PMID: 27539223 Free PMC article.
References
-
- Sirivatanauksorn V, Sirivatanauksorn Y, Lemoine NR. Molecular pattern of ductal pancreatic cancer. Langenbecks Arch Surg. 1998;383:105–115. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

