Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization
- PMID: 20117128
- PMCID: PMC2846990
- DOI: 10.1016/j.mad.2010.01.006
Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization
Abstract
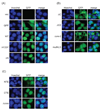
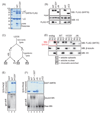
The mammalian sirtuin SIRT6 is a site-specific histone deacetylase that regulates chromatin structure. SIRT6 is implicated in fundamental biological processes in aging, including maintaining telomere integrity, fine-tuning aging-associated gene expression programs, preventing genomic instability, and maintaining metabolic homeostasis. Despite these important functions, the basic molecular determinants of SIRT6 enzymatic function--including the mechanistic and regulatory roles of specific domains of SIRT6--are not well understood. Sirtuin proteins consist of a conserved central 'sirtuin domain'--thought to comprise an enzymatic core--flanked by variable N- and C-terminal extensions. Here, we report the identification of novel functions for the N- and C-terminal domains of the human SIRT6 protein. We show that the C-terminal extension (CTE) of SIRT6 contributes to proper nuclear localization but is dispensable for enzymatic activity. In contrast, the N-terminal extension (NTE) of SIRT6 is critical for chromatin association and intrinsic catalytic activity. Surprisingly, mutation of a conserved catalytic histidine residue in the core sirtuin domain not only abrogates SIRT6 enzymatic activity but also leads to impaired chromatin association in cells. Together, our observations define important biochemical and cellular roles of specific SIRT6 domains, and provide mechanistic insight into the potential role of these domains as targets for physiologic and pharmacologic modulation.
Published by Elsevier Ireland Ltd.
Figures


Similar articles
-
Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators.J Biol Chem. 2020 Aug 7;295(32):11021-11041. doi: 10.1074/jbc.REV120.011438. Epub 2020 Jun 9. J Biol Chem. 2020. PMID: 32518153 Free PMC article. Review.
-
SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin.Nature. 2008 Mar 27;452(7186):492-6. doi: 10.1038/nature06736. Epub 2008 Mar 12. Nature. 2008. PMID: 18337721 Free PMC article.
-
Sirtuin 6: a review of biological effects and potential therapeutic properties.Mol Biosyst. 2013 Jul;9(7):1789-806. doi: 10.1039/c3mb00001j. Epub 2013 Apr 17. Mol Biosyst. 2013. PMID: 23592245 Review.
-
Chromatin regulation and genome maintenance by mammalian SIRT6.Trends Biochem Sci. 2011 Jan;36(1):39-46. doi: 10.1016/j.tibs.2010.07.009. Epub 2010 Aug 21. Trends Biochem Sci. 2011. PMID: 20729089 Free PMC article. Review.
-
SIRT6 facilitates directional telomere movement upon oxidative damage.Sci Rep. 2018 Mar 29;8(1):5407. doi: 10.1038/s41598-018-23602-0. Sci Rep. 2018. PMID: 29599436 Free PMC article.
Cited by
-
Activation of SIRT6 Deacetylation by DNA Strand Breaks.ACS Omega. 2023 Oct 23;8(44):41310-41320. doi: 10.1021/acsomega.3c04859. eCollection 2023 Nov 7. ACS Omega. 2023. PMID: 37970049 Free PMC article.
-
Structure and biochemical functions of SIRT6.J Biol Chem. 2011 Apr 22;286(16):14575-87. doi: 10.1074/jbc.M111.218990. Epub 2011 Mar 1. J Biol Chem. 2011. PMID: 21362626 Free PMC article.
-
Biological and catalytic functions of sirtuin 6 as targets for small-molecule modulators.J Biol Chem. 2020 Aug 7;295(32):11021-11041. doi: 10.1074/jbc.REV120.011438. Epub 2020 Jun 9. J Biol Chem. 2020. PMID: 32518153 Free PMC article. Review.
-
Pathways for ischemic cytoprotection: role of sirtuins in caloric restriction, resveratrol, and ischemic preconditioning.J Cereb Blood Flow Metab. 2011 Apr;31(4):1003-19. doi: 10.1038/jcbfm.2010.229. Epub 2011 Jan 12. J Cereb Blood Flow Metab. 2011. PMID: 21224864 Free PMC article. Review.
-
Role of Sirtuins in Linking Metabolic Syndrome with Depression.Front Cell Neurosci. 2016 Mar 31;10:86. doi: 10.3389/fncel.2016.00086. eCollection 2016. Front Cell Neurosci. 2016. PMID: 27065808 Free PMC article. Review.
References
-
- Denu JM. Linking chromatin function with metabolic networks: Sir2 family of NAD(+)-dependent deacetylases. Trends Biochem Sci. 2003;28:41–48. - PubMed
-
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. - PubMed
-
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem Biophys Res Commun. 2000;273:793–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

