Identification of the early VIP-regulated transcriptome and its associated, interactome in resting and activated murine CD4 T cells
- PMID: 20117839
- PMCID: PMC2834953
- DOI: 10.1016/j.molimm.2010.01.003
Identification of the early VIP-regulated transcriptome and its associated, interactome in resting and activated murine CD4 T cells
Abstract
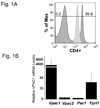
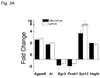
More than 40 years after the discovery of vasoactive intestinal peptide (VIP), its transcriptome in the immune system has still not been completely elucidated. In an attempt to understand the biological role of this neuropeptide in immunity, we chose CD4 T cells as a cellular system. Agilent Mouse Whole Genome microarrays were hybridized with fluorescently labeled total RNA isolated from resting CD4 T cells cultured +/-10(-7)M VIP for 5h or PMA/ionomycin activated CD4 T cells cultured +/-10(-7)M VIP for 5h. These VIP-regulated transcriptomes were analyzed by Significance Analysis of Microarrays (SAM) and Ingenuity Pathway Analysis (IPA) software to identify relevant signaling pathways modulated by VIP in the absence and presence of T cell activation. In resting CD4 T cells, VIP-modulated 368 genes, ranging from 3.49 to -4.78-fold. In the PMA/ionomycin activated CD4 T cells, 326 gene expression levels were changed by VIP, ranging from 2.94 to -1.66-fold. IPA analysis revealed that VIP exposure alters cellular function through EGFR signaling in resting CD4 T cells, and modulates immediate early genes, Fos and CREM/ICER, in activated CD4 T cells. These gene expression changes are suggested to explain at a molecular level how VIP can regulate T cell homing to the gut and induce regulatory T cell generation.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures








Similar articles
-
Selective gene expression and activation-dependent regulation of vasoactive intestinal peptide receptor type 1 and type 2 in human T cells.J Immunol. 2001 Feb 15;166(4):2522-30. doi: 10.4049/jimmunol.166.4.2522. J Immunol. 2001. PMID: 11160313
-
Roles of vasoactive intestinal peptide (VIP) in the expression of different immune phenotypes by wild-type mice and T cell-targeted type II VIP receptor transgenic mice.J Immunol. 2003 Jan 1;170(1):308-14. doi: 10.4049/jimmunol.170.1.308. J Immunol. 2003. PMID: 12496414
-
Effect of vasoactive intestinal peptide (VIP) on cytokine production and expression of VIP receptors in thymocyte subsets.Regul Pept. 1997 Sep 26;72(1):41-54. doi: 10.1016/s0167-0115(97)01028-8. Regul Pept. 1997. PMID: 9404731
-
Vasoactive intestinal peptide: a neuropeptide with pleiotropic immune functions.Amino Acids. 2013 Jul;45(1):25-39. doi: 10.1007/s00726-011-1184-8. Epub 2011 Dec 3. Amino Acids. 2013. PMID: 22139413 Free PMC article. Review.
-
Role of vasoactive intestinal peptide in inflammation and autoimmunity.Curr Opin Investig Drugs. 2005 Nov;6(11):1116-23. Curr Opin Investig Drugs. 2005. PMID: 16312132 Review.
Cited by
-
VPAC1 receptor (Vipr1)-deficient mice exhibit ameliorated experimental autoimmune encephalomyelitis, with specific deficits in the effector stage.J Neuroinflammation. 2016 Jun 29;13(1):169. doi: 10.1186/s12974-016-0626-3. J Neuroinflammation. 2016. PMID: 27357191 Free PMC article.
-
Radical reversal of vasoactive intestinal peptide (VIP) receptors during early lymphopoiesis.Peptides. 2011 Oct;32(10):2058-66. doi: 10.1016/j.peptides.2011.08.014. Epub 2011 Aug 22. Peptides. 2011. PMID: 21878358 Free PMC article.
-
Gene expression profiling in gastric mucosa from Helicobacter pylori-infected and uninfected patients undergoing chronic superficial gastritis.PLoS One. 2012;7(3):e33030. doi: 10.1371/journal.pone.0033030. Epub 2012 Mar 16. PLoS One. 2012. PMID: 22438889 Free PMC article.
-
Gene expression profiling and network analysis of peripheral blood monocytes in a chronic model of allergic asthma.Microbiol Immunol. 2010 Sep;54(9):558-63. doi: 10.1111/j.1348-0421.2010.00242.x. Microbiol Immunol. 2010. PMID: 20840155 Free PMC article.
-
Characterization and use of a rabbit-anti-mouse VPAC1 antibody by flow cytometry.J Immunol Methods. 2012 Feb 28;376(1-2):20-31. doi: 10.1016/j.jim.2011.10.009. Epub 2011 Nov 4. J Immunol Methods. 2012. PMID: 22079255 Free PMC article.
References
-
- Abad C, Juarranz Y, Martinez C, Arranz A, Rosignoli F, Garcia-Gomez M, Leceta J, Gomariz RP. cDNA array analysis of cytokines, chemokines, and receptors involved in the development of TNBS-induced colitis: homeostatic role of VIP. Inflamm Bowel Dis. 2005;11:674–684. - PubMed
-
- Abad C, Martinez C, Juarranz MG, Arranz A, Leceta J, Delgado M, Gomariz RP. Therapeutic effects of vasoactive intestinal peptide in the trinitrobenzene sulfonic acid mice model of Crohn's disease. Gastroenterology. 2003;124:961–971. - PubMed
-
- Arranz A, Gutierrez-Canas I, Carrion M, Juarranz Y, Pablos JL, Martinez C, Gomariz RP. VIP reverses the expression profiling of TLR4-stimulated signaling pathway in rheumatoid arthritis synovial fibroblasts. Mol Immunol. 2008;45:3065–3073. - PubMed
-
- Ben-Horin S, Chowers Y. Neuroimmunology of the gut: physiology, pathology, and pharmacology. Curr Opin Pharmacol. 2008;8:490–495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

