Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae)
- PMID: 20118192
- PMCID: PMC2877997
- DOI: 10.1093/molbev/msq029
Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae)
Abstract
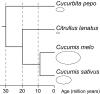
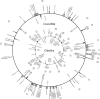
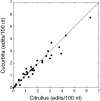
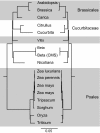
The mitochondrial genomes of seed plants are unusually large and vary in size by at least an order of magnitude. Much of this variation occurs within a single family, the Cucurbitaceae, whose genomes range from an estimated 390 to 2,900 kb in size. We sequenced the mitochondrial genomes of Citrullus lanatus (watermelon: 379,236 nt) and Cucurbita pepo (zucchini: 982,833 nt)--the two smallest characterized cucurbit mitochondrial genomes--and determined their RNA editing content. The relatively compact Citrullus mitochondrial genome actually contains more and longer genes and introns, longer segmental duplications, and more discernibly nuclear-derived DNA. The large size of the Cucurbita mitochondrial genome reflects the accumulation of unprecedented amounts of both chloroplast sequences (>113 kb) and short repeated sequences (>370 kb). A low mutation rate has been hypothesized to underlie increases in both genome size and RNA editing frequency in plant mitochondria. However, despite its much larger genome, Cucurbita has a significantly higher synonymous substitution rate (and presumably mutation rate) than Citrullus but comparable levels of RNA editing. The evolution of mutation rate, genome size, and RNA editing are apparently decoupled in Cucurbitaceae, reflecting either simple stochastic variation or governance by different factors.
Figures
References
-
- André C, Levy A, Walbot V. Small repeated sequences and the structure of plant mitochondrial genomes. Trends Genet. 1992;8:128–132. - PubMed
-
- Barker NP, Clark LG, Davis JI, et al. 13 co-authors. Phylogeny and subfamilial classification of the grasses (Poaceae) Ann Mo Bot Gard. 2001;88:373–457.
-
- Bartoszewski G, Gawronski P, Szklarczyk M, Verbakel H, Havey MJ. A one-megabase physical map provides insights on gene organization in the enormous mitochondrial genome of cucumber. Genome. 2009;52:299–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

