Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis
- PMID: 20118213
- PMCID: PMC2837984
- DOI: 10.1128/EC.00358-09
Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis
Abstract
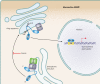
Sterol regulatory element binding proteins (SREBPs) are membrane-bound transcription factors whose proteolytic activation is controlled by the cellular sterol concentration. Mammalian SREBPs are activated in cholesterol-depleted cells and serve to regulate cellular lipid homeostasis. Recent work demonstrates that SREBP is functionally conserved in fungi. While the ability to respond to sterols is conserved, fungal SREBPs are hypoxic transcription factors required for adaptation to a low-oxygen environment. In the fission yeast Schizosaccharomyces pombe, oxygen regulates the SREBP homolog Sre1 by independently controlling both its proteolytic activation and its degradation. SREBP is also required for adaptation to hypoxia in the human pathogens Cryptococcus neoformans and Aspergillus fumigatus. In these organisms, SREBP is required for virulence and resistance to antifungal drugs, making the SREBP pathway a potential target for antifungal therapy.
Figures
References
-
- Brown A. J., Sun L. P., Feramisco J. D., Brown M. S., Goldstein J. L. 2002. Cholesterol addition to ER membranes alters conformation of SCAP, the SREBP escort protein that regulates cholesterol metabolism. Mol. Cell 10:237–245 - PubMed
-
- Brown M. S., Goldstein J. L. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331–340 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

