Evolutionary studies illuminate the structural-functional model of plant phytochromes
- PMID: 20118225
- PMCID: PMC2828699
- DOI: 10.1105/tpc.109.072280
Evolutionary studies illuminate the structural-functional model of plant phytochromes
Abstract
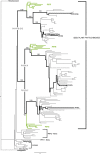
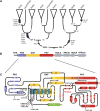
A synthesis of insights from functional and evolutionary studies reveals how the phytochrome photoreceptor system has evolved to impart both stability and flexibility. Phytochromes in seed plants diverged into three major forms, phyA, phyB, and phyC, very early in the history of seed plants. Two additional forms, phyE and phyD, are restricted to flowering plants and Brassicaceae, respectively. While phyC, D, and E are absent from at least some taxa, phyA and phyB are present in all sampled seed plants and are the principal mediators of red/far-red-induced responses. Conversely, phyC-E apparently function in concert with phyB and, where present, expand the repertoire of phyB activities. Despite major advances, aspects of the structural-functional models for these photoreceptors remain elusive. Comparative sequence analyses expand the array of locus-specific mutant alleles for analysis by revealing historic mutations that occurred during gene lineage splitting and divergence. With insights from crystallographic data, a subset of these mutants can be chosen for functional studies to test their importance and determine the molecular mechanism by which they might impact light perception and signaling. In the case of gene families, where redundancy hinders isolation of some proportion of the relevant mutants, the approach may be particularly useful.
Figures
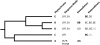
Similar articles
-
Evolutionary aspects of plant photoreceptors.J Plant Res. 2016 Mar;129(2):115-22. doi: 10.1007/s10265-016-0785-4. Epub 2016 Feb 3. J Plant Res. 2016. PMID: 26843269 Review.
-
Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways.Plant Cell. 2003 Sep;15(9):1962-80. doi: 10.1105/tpc.012971. Plant Cell. 2003. PMID: 12953104 Free PMC article.
-
Duplication, divergence and persistence in the Phytochrome photoreceptor gene family of cottons (Gossypium spp.).BMC Plant Biol. 2010 Jun 20;10:119. doi: 10.1186/1471-2229-10-119. BMC Plant Biol. 2010. PMID: 20565911 Free PMC article.
-
Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice.Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14705-10. doi: 10.1073/pnas.0907378106. Epub 2009 Aug 12. Proc Natl Acad Sci U S A. 2009. PMID: 19706555 Free PMC article.
-
The system of phytochromes: photobiophysics and photobiochemistry in vivo.Membr Cell Biol. 1998;12(5):691-720. Membr Cell Biol. 1998. PMID: 10379648 Review.
Cited by
-
Conformational Change of Tetratricopeptide Repeats Region Triggers Activation of Phytochrome-Associated Protein Phosphatase 5.Front Plant Sci. 2021 Oct 14;12:733069. doi: 10.3389/fpls.2021.733069. eCollection 2021. Front Plant Sci. 2021. PMID: 34721460 Free PMC article.
-
Photosensing and Thermosensing by Phytochrome B Require Both Proximal and Distal Allosteric Features within the Dimeric Photoreceptor.Sci Rep. 2017 Oct 20;7(1):13648. doi: 10.1038/s41598-017-14037-0. Sci Rep. 2017. PMID: 29057954 Free PMC article.
-
Evolutionary aspects of plant photoreceptors.J Plant Res. 2016 Mar;129(2):115-22. doi: 10.1007/s10265-016-0785-4. Epub 2016 Feb 3. J Plant Res. 2016. PMID: 26843269 Review.
-
Regulation of monocot and dicot plant development with constitutively active alleles of phytochrome B.Plant Direct. 2020 Apr 27;4(4):e00210. doi: 10.1002/pld3.210. eCollection 2020 Apr. Plant Direct. 2020. PMID: 32346668 Free PMC article.
-
Molecular mechanisms underlying phytochrome-controlled morphogenesis in plants.Nat Commun. 2019 Nov 19;10(1):5219. doi: 10.1038/s41467-019-13045-0. Nat Commun. 2019. PMID: 31745087 Free PMC article. Review.
References
-
- Alba R., Kelmenson P.M., Cordonnier-Pratt M.M., Pratt L.H. (2000). The phytochrome gene family in tomato and the rapid differential evolution of this family in angiosperms. Mol. Biol. Evol. 17: 362–373 - PubMed
-
- Alonso-Blanco C., Koornneef M. (2000). Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 5: 22–29 - PubMed
-
- Bae G., Choi G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59: 281–311 - PubMed
-
- Balasubramanian S., Sureshkumar S., Agrawal M., Michael T.P., Wessinger C., Maloof J.N., Clark R., Warthmann N., Chory J., Weigel D. (2006). The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nat. Genet. 38: 711–715 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

