The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana
- PMID: 20118226
- PMCID: PMC2828693
- DOI: 10.1105/tpc.109.071803
The ABORTED MICROSPORES regulatory network is required for postmeiotic male reproductive development in Arabidopsis thaliana
Abstract
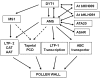
The Arabidopsis thaliana ABORTED MICROSPORES (AMS) gene encodes a basic helix-loop-helix (bHLH) transcription factor that is required for tapetal cell development and postmeiotic microspore formation. However, the regulatory role of AMS in anther and pollen development has not been fully defined. Here, we show by microarray analysis that the expression of 549 anther-expressed genes was altered in ams buds and that these genes are associated with tapetal function and pollen wall formation. We demonstrate that AMS has the ability to bind in vitro to DNA containing a 6-bp consensus motif, CANNTG. Moreover, 13 genes involved in transportation of lipids, oligopeptides, and ions, fatty acid synthesis and metabolism, flavonol accumulation, substrate oxidation, methyl-modification, and pectin dynamics were identified as direct targets of AMS by chromatin immunoprecipitation. The functional importance of the AMS regulatory pathway was further demonstrated by analysis of an insertional mutant of one of these downstream AMS targets, an ABC transporter, White-Brown Complex homolog, which fails to undergo pollen development and is male sterile. Yeast two-hybrid screens and pull-down assays revealed that AMS has the ability to interact with two bHLH proteins (AtbHLH089 and AtbHLH091) and the ATA20 protein. These results provide insight into the regulatory role of the AMS network during anther development.
Figures





Similar articles
-
Biphasic regulation of the transcription factor ABORTED MICROSPORES (AMS) is essential for tapetum and pollen development in Arabidopsis.New Phytol. 2017 Jan;213(2):778-790. doi: 10.1111/nph.14200. Epub 2016 Oct 27. New Phytol. 2017. PMID: 27787905 Free PMC article.
-
AMS-dependent and independent regulation of anther transcriptome and comparison with those affected by other Arabidopsis anther genes.BMC Plant Biol. 2012 Feb 15;12:23. doi: 10.1186/1471-2229-12-23. BMC Plant Biol. 2012. PMID: 22336428 Free PMC article.
-
Positive regulation of AMS by TDF1 and the formation of a TDF1-AMS complex are required for anther development in Arabidopsis thaliana.New Phytol. 2018 Jan;217(1):378-391. doi: 10.1111/nph.14790. Epub 2017 Sep 20. New Phytol. 2018. PMID: 28940573
-
DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis.Plant J. 2014 Dec;80(6):1005-13. doi: 10.1111/tpj.12694. Epub 2014 Nov 6. Plant J. 2014. PMID: 25284309
-
Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis.Int J Mol Sci. 2021 Jul 1;22(13):7152. doi: 10.3390/ijms22137152. Int J Mol Sci. 2021. PMID: 34281206 Free PMC article. Review.
Cited by
-
Comparative Transcriptome Analysis Reveals Key Insights into Fertility Conversion in the Thermo-Sensitive Cytoplasmic Male Sterile Wheat.Int J Mol Sci. 2022 Nov 18;23(22):14354. doi: 10.3390/ijms232214354. Int J Mol Sci. 2022. PMID: 36430832 Free PMC article.
-
Global transcriptome analysis reveals potential genes associated with genic male sterility of rapeseed (Brassica napus L.).Front Plant Sci. 2022 Oct 21;13:1004781. doi: 10.3389/fpls.2022.1004781. eCollection 2022. Front Plant Sci. 2022. PMID: 36340380 Free PMC article.
-
A regulatory network for coordinated flower maturation.PLoS Genet. 2012 Feb;8(2):e1002506. doi: 10.1371/journal.pgen.1002506. Epub 2012 Feb 9. PLoS Genet. 2012. PMID: 22346763 Free PMC article.
-
Transcriptomic analysis reveals candidate genes associated with anther development in Lilium Oriental Hybrid 'Siberia'.Front Plant Sci. 2023 Feb 8;14:1128911. doi: 10.3389/fpls.2023.1128911. eCollection 2023. Front Plant Sci. 2023. PMID: 36844086 Free PMC article.
-
Exon junction complex (EJC) core genes play multiple developmental roles in Physalis floridana.Plant Mol Biol. 2018 Dec;98(6):545-563. doi: 10.1007/s11103-018-0795-9. Epub 2018 Nov 13. Plant Mol Biol. 2018. PMID: 30426309 Free PMC article.
References
-
- Aarts M.G., Hodge R., Kalantidis K., Florack D., Wilson Z.A., Mulligan B.J., Stiekema W.J., Scott R., Pereira A. (1997). The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 12: 615–623 - PubMed
-
- Ageez A., Kazama Y., Sugiyama R., Kawano S. (2005). Male-fertility genes expressed in male flower buds of Silene latifolia include homologs of anther-specific genes. Genes Genet. Syst. 80: 403–413 - PubMed
-
- Alexander M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44: 117–122 - PubMed
-
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

