G9a and Glp methylate lysine 373 in the tumor suppressor p53
- PMID: 20118233
- PMCID: PMC2843213
- DOI: 10.1074/jbc.M109.062588
G9a and Glp methylate lysine 373 in the tumor suppressor p53
Erratum in
- J Biol Chem. 2010 Jun 4;285(23):18122. Pérez-Burgos, Laura [added]
Abstract
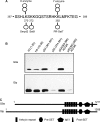
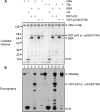
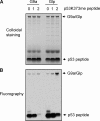
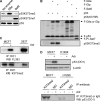
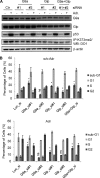
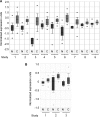
The tumor suppressor p53 is regulated by numerous post-translational modifications. Lysine methylation has recently emerged as a key post-translational modification that alters the activity of p53. Here, we describe a novel lysine methylation site in p53 that is carried out by two homologous histone methyltransferases, G9a and Glp. G9a and Glp specifically methylate p53 at Lys(373), resulting mainly in dimethylation. During DNA damage, the overall level of p53 modified at Lys(373)me2 does not increase, despite the dramatic increase in total p53, indicating that Lys(373)me2 correlates with inactive p53. Further, reduction of G9a and/or Glp levels leads to a larger population of apoptotic cells. Examination of the Oncomine data base shows that G9a and Glp are overexpressed in various cancers compared with corresponding normal tissues, suggesting that they are putative oncogenes. These data reveal a new methylation site within p53 mediated by the methylases G9a and Glp and indicate that G9a is a potential inhibitory target for cancer treatment.
Figures
References
-
- Bode A. M., Dong Z. (2004) Nat. Rev. Cancer 4, 793–805 - PubMed
-
- Prives C., Manley J. L. (2001) Cell 107, 815–818 - PubMed
-
- Huang J., Berger S. L. (2008) Curr. Opin. Genet. Dev. 18, 152–158 - PubMed
-
- Huang J., Perez-Burgos L., Placek B. J., Sengupta R., Richter M., Dorsey J. A., Lysubicek S., Opravil S., Jenuwein T., Berger S. L. (2006) Nature 444, 629–632 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

