A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity
- PMID: 20118235
- PMCID: PMC2856252
- DOI: 10.1074/jbc.M109.093427
A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity
Abstract
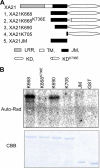
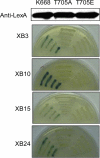
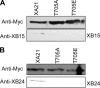
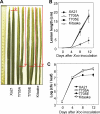
Despite the key role that pattern recognition receptors (PRRs) play in regulating immunity in plants and animals, the mechanism of activation of the associated non-arginine-aspartate (non-RD) kinases is unknown. The rice PRR XA21 recognizes the pathogen-associated molecular pattern, Ax21 (activator of XA21-mediated immunity). Here we show that the XA21 juxtamembrane (JM) domain is required for kinase autophosphorylation. Threonine 705 in the XA21 JM domain is essential for XA21 autophosphorylation in vitro and XA21-mediated innate immunity in vivo. The replacement of Thr(705) by an alanine or glutamic acid abolishes XA21 autophosphorylation and eliminates interactions between XA21 and four XA21-binding proteins in yeast and rice. Although threonine residues analogous to Thr(705) of XA21 are present in the JM domains of most RD and non-RD plant receptor-like kinases, this residue is not required for autophosphorylation of the Arabidopsis RD RLK BRI1 (brassinosteroid insensitive 1). The threonine 705 of XA21 is conserved only in the JM domains of plant RLKs but not in those of fly, human, or mouse suggesting distinct regulatory mechanisms. These results contribute to growing knowledge regarding the mechanism by which non-RD RLKs function in plant.
Figures





References
-
- Wang X., Li X., Meisenhelder J., Hunter T., Yoshida S., Asami T., Chory J. (2005) Dev. Cell 8, 855–865 - PubMed
-
- Heiss E., Masson K., Sundberg C., Pedersen M., Sun J., Bengtsson S., Rönnstrand L. (2006) Blood 108, 1542–1550 - PubMed
-
- Aifa S., Frikha F., Miled N., Johansen K., Lundström I., Svensson S. P. (2006) Biochem. Biophys. Res. Commun. 347, 381–387 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

