Role for CysJ flavin reductase in molybdenum cofactor-dependent resistance of Escherichia coli to 6-N-hydroxylaminopurine
- PMID: 20118259
- PMCID: PMC2849459
- DOI: 10.1128/JB.01438-09
Role for CysJ flavin reductase in molybdenum cofactor-dependent resistance of Escherichia coli to 6-N-hydroxylaminopurine
Abstract
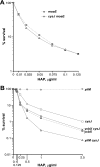
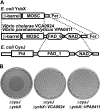
We have previously described a novel Escherichia coli detoxification system for the removal of toxic and mutagenic N-hydroxylated nucleobases and related compounds that requires the molybdenum cofactor. Two subpathways (ycbX and yiiM) were identified, each employing a novel molybdo activity capable of inactivating N-hydroxylated compounds by reduction to the corresponding amine. In the present study, we identify the cysJ gene product as one additional component of this system. While the CysJ protein has been identified as the NADPH:flavin oxidoreductase component of the CysJI sulfite reductase complex (CysJ(8)I(4)), we show that the role of CysJ in base analog detoxification is unique and independent of CysI and sulfite reductase. We further show that CysJ functions as a specific partner of the YcbX molybdoenzyme. We postulate that the function of CysJ in this pathway is to provide, via its NADPH:flavin reductase activity, the reducing equivalents needed for the detoxification reaction at the YcbX molybdocenter. In support of the proposed interaction of the CysJ and YcbX proteins, we show that an apparent CysJ-YcbX "hybrid" protein from two Vibrio species is capable of compensating for a double cysJ ycbX defect in E. coli.
Figures
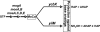




Comment in
-
The twists and turns of enzyme function.J Bacteriol. 2010 Apr;192(8):2023-5. doi: 10.1128/JB.00087-10. Epub 2010 Feb 12. J Bacteriol. 2010. PMID: 20154124 Free PMC article. No abstract available.
References
-
- Anantharaman, V., and L. Aravind. 2002. MOSC domains: ancient, predicted sulfur-carrier domains, present in diverse metal-sulfur cluster biosynthesis proteins including molybdenum cofactor sulfurases. FEMS Microbiol. Lett. 207:55-61. - PubMed
-
- Butland, G., J. M. Peregrín-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. - PubMed
-
- Clement, B., and T. Kunze. 1990. Hepatic microsomal N-hydroxylation of adenine to 6-N-hydroxylaminopurine. Biochem. Pharmacol. 39:925-933. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

