Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor
- PMID: 20118260
- PMCID: PMC2838060
- DOI: 10.1128/JB.01348-09
Crystal structure of the cystic fibrosis transmembrane conductance regulator inhibitory factor Cif reveals novel active-site features of an epoxide hydrolase virulence factor
Abstract
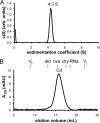
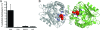
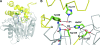
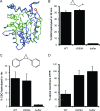
Cystic fibrosis transmembrane conductance regulator (CFTR) inhibitory factor (Cif) is a virulence factor secreted by Pseudomonas aeruginosa that reduces the quantity of CFTR in the apical membrane of human airway epithelial cells. Initial sequence analysis suggested that Cif is an epoxide hydrolase (EH), but its sequence violates two strictly conserved EH motifs and also is compatible with other alpha/beta hydrolase family members with diverse substrate specificities. To investigate the mechanistic basis of Cif activity, we have determined its structure at 1.8-A resolution by X-ray crystallography. The catalytic triad consists of residues Asp129, His297, and Glu153, which are conserved across the family of EHs. At other positions, sequence deviations from canonical EH active-site motifs are stereochemically conservative. Furthermore, detailed enzymatic analysis confirms that Cif catalyzes the hydrolysis of epoxide compounds, with specific activity against both epibromohydrin and cis-stilbene oxide, but with a relatively narrow range of substrate selectivity. Although closely related to two other classes of alpha/beta hydrolase in both sequence and structure, Cif does not exhibit activity as either a haloacetate dehalogenase or a haloalkane dehalogenase. A reassessment of the structural and functional consequences of the H269A mutation suggests that Cif's effect on host-cell CFTR expression requires the hydrolysis of an extended endogenous epoxide substrate.
Figures
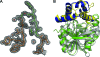

Similar articles
-
Pouring salt on a wound: Pseudomonas aeruginosa virulence factors alter Na+ and Cl- flux in the lung.J Bacteriol. 2013 Sep;195(18):4013-9. doi: 10.1128/JB.00339-13. Epub 2013 Jul 8. J Bacteriol. 2013. PMID: 23836869 Free PMC article. Review.
-
Pseudomonas aeruginosa Cif defines a distinct class of α/β epoxide hydrolases utilizing a His/Tyr ring-opening pair.Protein Pept Lett. 2012 Feb;19(2):186-93. doi: 10.2174/092986612799080392. Protein Pept Lett. 2012. PMID: 21933119 Free PMC article.
-
Signature motifs identify an Acinetobacter Cif virulence factor with epoxide hydrolase activity.J Biol Chem. 2014 Mar 14;289(11):7460-9. doi: 10.1074/jbc.M113.518092. Epub 2014 Jan 28. J Biol Chem. 2014. PMID: 24474692 Free PMC article.
-
Active-Site Flexibility and Substrate Specificity in a Bacterial Virulence Factor: Crystallographic Snapshots of an Epoxide Hydrolase.Structure. 2017 May 2;25(5):697-707.e4. doi: 10.1016/j.str.2017.03.002. Epub 2017 Apr 6. Structure. 2017. PMID: 28392259 Free PMC article.
-
The telltale structures of epoxide hydrolases.Drug Metab Rev. 2003 Nov;35(4):365-83. doi: 10.1081/dmr-120026498. Drug Metab Rev. 2003. PMID: 14705866 Review.
Cited by
-
Visualizing the Mechanism of Epoxide Hydrolysis by the Bacterial Virulence Enzyme Cif.Biochemistry. 2016 Feb 9;55(5):788-97. doi: 10.1021/acs.biochem.5b01229. Epub 2016 Jan 22. Biochemistry. 2016. PMID: 26752215 Free PMC article.
-
Pouring salt on a wound: Pseudomonas aeruginosa virulence factors alter Na+ and Cl- flux in the lung.J Bacteriol. 2013 Sep;195(18):4013-9. doi: 10.1128/JB.00339-13. Epub 2013 Jul 8. J Bacteriol. 2013. PMID: 23836869 Free PMC article. Review.
-
Pseudomonas aeruginosa Cif defines a distinct class of α/β epoxide hydrolases utilizing a His/Tyr ring-opening pair.Protein Pept Lett. 2012 Feb;19(2):186-93. doi: 10.2174/092986612799080392. Protein Pept Lett. 2012. PMID: 21933119 Free PMC article.
-
Exported Epoxide Hydrolases Modulate Erythrocyte Vasoactive Lipids during Plasmodium falciparum Infection.mBio. 2016 Oct 18;7(5):e01538-16. doi: 10.1128/mBio.01538-16. mBio. 2016. PMID: 27795395 Free PMC article.
-
Inhibiting an Epoxide Hydrolase Virulence Factor from Pseudomonas aeruginosa Protects CFTR.Angew Chem Int Ed Engl. 2015 Aug 17;54(34):9881-5. doi: 10.1002/anie.201503983. Epub 2015 Jul 1. Angew Chem Int Ed Engl. 2015. PMID: 26136396 Free PMC article.
References
-
- American Thoracic Society and Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am. J. Respir. Crit. Care Med. 171:388-416. - PubMed
-
- Arand, M., A. Cronin, M. Adamska, and F. Oesch. 2005. Epoxide hydrolases: structure, function, mechanism, and assay. Methods Enzymol. 400:569-588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

