Sequence analysis of Leuconostoc mesenteroides bacteriophage Phi1-A4 isolated from an industrial vegetable fermentation
- PMID: 20118355
- PMCID: PMC2838018
- DOI: 10.1128/AEM.02126-09
Sequence analysis of Leuconostoc mesenteroides bacteriophage Phi1-A4 isolated from an industrial vegetable fermentation
Abstract
Vegetable fermentations rely on the proper succession of a variety of lactic acid bacteria (LAB). Leuconostoc mesenteroides initiates fermentation. As fermentation proceeds, L. mesenteroides dies off and other LAB complete the fermentation. Phages infecting L. mesenteroides may significantly influence the die-off of L. mesenteroides. However, no L. mesenteroides phages have been previously genetically characterized. Knowledge of more phage genome sequences may provide new insights into phage genomics, phage evolution, and phage-host interactions. We have determined the complete genome sequence of L. mesenteroides phage Phi1-A4, isolated from an industrial sauerkraut fermentation. The phage possesses a linear, double-stranded DNA genome consisting of 29,508 bp with a G+C content of 36%. Fifty open reading frames (ORFs) were predicted. Putative functions were assigned to 26 ORFs (52%), including 5 ORFs of structural proteins. The phage genome was modularly organized, containing DNA replication, DNA-packaging, head and tail morphogenesis, cell lysis, and DNA regulation/modification modules. In silico analyses showed that Phi1-A4 is a unique lytic phage with a large-scale genome inversion ( approximately 30% of the genome). The genome inversion encompassed the lysis module, part of the structural protein module, and a cos site. The endolysin gene was flanked by two holin genes. The tail morphogenesis module was interspersed with cell lysis genes and other genes with unknown functions. The predicted amino acid sequences of the phage proteins showed little similarity to other phages, but functional analyses showed that Phi1-A4 clusters with several Lactococcus phages. To our knowledge, Phi1-A4 is the first genetically characterized L. mesenteroides phage.
Figures
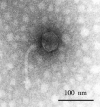





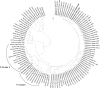
Similar articles
-
Phages of dairy Leuconostoc mesenteroides: genomics and factors influencing their adsorption.Int J Food Microbiol. 2015 May 18;201:58-65. doi: 10.1016/j.ijfoodmicro.2015.02.016. Epub 2015 Feb 21. Int J Food Microbiol. 2015. PMID: 25747109
-
Sequence and comparative analysis of Leuconostoc dairy bacteriophages.Int J Food Microbiol. 2014 Apr 17;176:29-37. doi: 10.1016/j.ijfoodmicro.2014.01.019. Epub 2014 Feb 5. Int J Food Microbiol. 2014. PMID: 24561391
-
Complete genome sequence of ΦMH1, a Leuconostoc temperate phage.Arch Virol. 2010 Nov;155(11):1883-5. doi: 10.1007/s00705-010-0799-5. Epub 2010 Sep 17. Arch Virol. 2010. PMID: 20848296
-
Genomes of the T4-related bacteriophages as windows on microbial genome evolution.Virol J. 2010 Oct 28;7:292. doi: 10.1186/1743-422X-7-292. Virol J. 2010. PMID: 21029436 Free PMC article. Review.
-
Bacteriophages of leuconostoc, oenococcus, and weissella.Front Microbiol. 2014 Apr 28;5:186. doi: 10.3389/fmicb.2014.00186. eCollection 2014. Front Microbiol. 2014. PMID: 24817864 Free PMC article. Review.
Cited by
-
Genome sequence and characterization of the Tsukamurella bacteriophage TPA2.Appl Environ Microbiol. 2011 Feb;77(4):1389-98. doi: 10.1128/AEM.01938-10. Epub 2010 Dec 23. Appl Environ Microbiol. 2011. PMID: 21183635 Free PMC article.
-
Identification of the receptor-binding protein in lytic Leuconostoc pseudomesenteroides bacteriophages.Appl Environ Microbiol. 2013 May;79(10):3311-4. doi: 10.1128/AEM.00012-13. Epub 2013 Mar 15. Appl Environ Microbiol. 2013. PMID: 23503306 Free PMC article.
-
Metagenomic analysis of the viral communities in fermented foods.Appl Environ Microbiol. 2011 Feb;77(4):1284-91. doi: 10.1128/AEM.01859-10. Epub 2010 Dec 23. Appl Environ Microbiol. 2011. PMID: 21183634 Free PMC article.
-
Tracing lifestyle adaptation in prokaryotic genomes.Front Microbiol. 2012 Feb 21;3:48. doi: 10.3389/fmicb.2012.00048. eCollection 2012. Front Microbiol. 2012. PMID: 22363326 Free PMC article.
-
Potential for Bacteriophage Endolysins to Supplement or Replace Antibiotics in Food Production and Clinical Care.Antibiotics (Basel). 2018 Feb 27;7(1):17. doi: 10.3390/antibiotics7010017. Antibiotics (Basel). 2018. PMID: 29495476 Free PMC article. Review.
References
-
- Acebo, P., M. Garcia de Lacoba, G. Rivas, J. M. Andreu, M. Espinosa, and G. del Solar. 1998. Structural features of the plasmid pMV158-encoded transcriptional repressor CopG, a protein sharing similarities with both helix-turn-helix and beta-sheet DNA binding proteins. Proteins 32:248-261. - PubMed
-
- Altermann, E., and T. R. Klaenhammer. 2003. GAMOLA: a new local solution for sequence annotation and analyzing draft and finished prokaryotic genomes. OMICS 7:161-169. - PubMed
-
- Altermann, E., J. R. Klein, and B. Henrich. 1999. Primary structure and features of the genome of the Lactobacillus gasseri temperate bacteriophage (phi)adh. Gene 236:333-346. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

