Evolutionary dynamics of clustered irregularly interspaced short palindromic repeat systems in the ocean metagenome
- PMID: 20118362
- PMCID: PMC2849227
- DOI: 10.1128/AEM.01985-09
Evolutionary dynamics of clustered irregularly interspaced short palindromic repeat systems in the ocean metagenome
Abstract
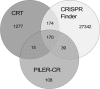
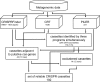
Clustered regularly interspaced short palindromic repeats (CRISPRs) form a recently characterized type of prokaryotic antiphage defense system. The phage-host interactions involving CRISPRs have been studied in experiments with selected bacterial or archaeal species and, computationally, in completely sequenced genomes. However, these studies do not allow one to take prokaryotic population diversity and phage-host interaction dynamics into account. This gap can be filled by using metagenomic data: in particular, the largest existing data set, generated from the Sorcerer II Global Ocean Sampling expedition. The application of three publicly available CRISPR recognition programs to the Global Ocean metagenome produced a large proportion of false-positive results. To address this problem, a filtering procedure was designed. It resulted in about 200 reliable CRISPR cassettes, which were then studied in detail. The repeat consensuses were clustered into several stable classes that differed from the existing classification. Short fragments of DNA similar to the cassette spacers were more frequently present in the same geographical location than in other locations (P, <0.0001). We developed a catalogue of elementary CRISPR-forming events and reconstructed the likely evolutionary history of cassettes that had common spacers. Metagenomic collections allow for relatively unbiased analysis of phage-host interactions and CRISPR evolution. The results of this study demonstrate that CRISPR cassettes retain the memory of the local virus population at a particular ocean location. CRISPR evolution may be described using a limited vocabulary of elementary events that have a natural biological interpretation.
Figures

References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Andersson, A. F., and J. F. Banfield. 2008. Virus population dynamics and acquired virus resistance in natural microbial communities. Science 320:1047-1050. - PubMed
-
- Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. - PMC - PubMed
-
- Arber, W. 1979. Promotion and limitation of genetic exchange. Science 205:361-365. - PubMed
-
- Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709-1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

