Gibberellic acid and cGMP-dependent transcriptional regulation in Arabidopsis thaliana
- PMID: 20118660
- PMCID: PMC2881265
- DOI: 10.4161/psb.5.3.10718
Gibberellic acid and cGMP-dependent transcriptional regulation in Arabidopsis thaliana
Abstract
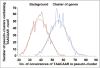
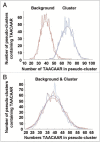
An ever increasing amount of transcriptomic data and analysis tools provide novel insight into complex responses of biological systems. Given these resources we have undertaken to review aspects of transcriptional regulation in response to the plant hormone gibberellic acid (GA) and its second messenger guanosine 3',5'-cyclic monophosphate (cGMP) in Arabidopsis thaliana, both wild type and selected mutants. Evidence suggests enrichment of GA-responsive (GARE) elements in promoters of genes that are transcriptionally upregulated in response to cGMP but downregulated in a GA insensitive mutant (ga1-3). In contrast, in the genes upregulated in the mutant, no enrichment in the GARE is observed suggesting that GARE motifs are diagnostic for GA-induced and cGMP-dependent transcriptional upregulation. Further, we review how expression studies of GA-dependent transcription factors and transcriptional networks based on common promoter signatures derived from ab initio analyses can contribute to our understanding of plant responses at the systems level.
Figures



Similar articles
-
BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin-mediated responses in arabidopsis.PLoS One. 2013;8(3):e58588. doi: 10.1371/journal.pone.0058588. Epub 2013 Mar 11. PLoS One. 2013. PMID: 23536800 Free PMC article.
-
Cyclic GMP is involved in auxin signalling during Arabidopsis root growth and development.J Exp Bot. 2014 Apr;65(6):1571-83. doi: 10.1093/jxb/eru019. Epub 2014 Mar 3. J Exp Bot. 2014. PMID: 24591051 Free PMC article.
-
SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response.Plant Physiol. 2011 Dec;157(4):1900-13. doi: 10.1104/pp.111.187302. Epub 2011 Oct 19. Plant Physiol. 2011. PMID: 22013217 Free PMC article.
-
Genomic analysis of DELLA protein activity.Plant Cell Physiol. 2013 Aug;54(8):1229-37. doi: 10.1093/pcp/pct082. Epub 2013 Jun 18. Plant Cell Physiol. 2013. PMID: 23784221 Review.
-
Gibberellin receptor and its role in gibberellin signaling in plants.Annu Rev Plant Biol. 2007;58:183-98. doi: 10.1146/annurev.arplant.58.032806.103830. Annu Rev Plant Biol. 2007. PMID: 17472566 Review.
Cited by
-
Genome-Wide Identification, Transcript Profiling and Bioinformatic Analyses of GRAS Transcription Factor Genes in Rice.Front Plant Sci. 2021 Nov 26;12:777285. doi: 10.3389/fpls.2021.777285. eCollection 2021. Front Plant Sci. 2021. PMID: 34899804 Free PMC article.
-
BIIDXI, the At4g32460 DUF642 gene, is involved in pectin methyl esterase regulation during Arabidopsis thaliana seed germination and plant development.BMC Plant Biol. 2014 Dec 2;14:338. doi: 10.1186/s12870-014-0338-8. BMC Plant Biol. 2014. PMID: 25442819 Free PMC article.
-
The arabidopsis cyclic nucleotide interactome.Cell Commun Signal. 2016 May 11;14(1):10. doi: 10.1186/s12964-016-0133-2. Cell Commun Signal. 2016. PMID: 27170143 Free PMC article.
-
Elevated concentrations of soil carbon dioxide with partial root-zone drying enhance drought tolerance and agro-physiological characteristics by regulating the expression of genes related to aquaporin and stress response in cucumber plants.BMC Plant Biol. 2024 Oct 1;24(1):917. doi: 10.1186/s12870-024-05310-2. BMC Plant Biol. 2024. PMID: 39354350 Free PMC article.
-
Endogenous NO levels regulate nodule functioning: potential role of cGMP in nodule functioning?Plant Signal Behav. 2010 Dec;5(12):1679-81. doi: 10.4161/psb.5.12.14041. Epub 2010 Dec 1. Plant Signal Behav. 2010. PMID: 21150263 Free PMC article.
References
-
- Lindsey K. Plant peptide hormones: The long and the short of it. Curr Biol. 2001;11:741–743. - PubMed
-
- Schaller H. The role of sterols in plant growth and development. Prog Lipid Res. 2003;42:163–175. - PubMed
-
- Neill SJ, Desikan R, Hancock JT. Nitric oxide signalling in plants. New Phytol. 2003;159:11–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
