Retroviral intasome assembly and inhibition of DNA strand transfer
- PMID: 20118915
- PMCID: PMC2837123
- DOI: 10.1038/nature08784
Retroviral intasome assembly and inhibition of DNA strand transfer
Abstract
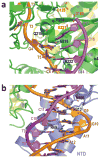
Integrase is an essential retroviral enzyme that binds both termini of linear viral DNA and inserts them into a host cell chromosome. The structure of full-length retroviral integrase, either separately or in complex with DNA, has been lacking. Furthermore, although clinically useful inhibitors of HIV integrase have been developed, their mechanism of action remains speculative. Here we present a crystal structure of full-length integrase from the prototype foamy virus in complex with its cognate DNA. The structure shows the organization of the retroviral intasome comprising an integrase tetramer tightly associated with a pair of viral DNA ends. All three canonical integrase structural domains are involved in extensive protein-DNA and protein-protein interactions. The binding of strand-transfer inhibitors displaces the reactive viral DNA end from the active site, disarming the viral nucleoprotein complex. Our findings define the structural basis of retroviral DNA integration, and will allow modelling of the HIV-1 intasome to aid in the development of antiretroviral drugs.
Figures
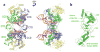


Comment in
-
Structural biology: When four become one.Nature. 2010 Mar 11;464(7286):167-8. doi: 10.1038/464167a. Nature. 2010. PMID: 20220826 Free PMC article.
References
-
- Craigie R. In: Mobile DNA II. Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. ASM Press; 2002. pp. 613–630.
-
- Lewinski MK, Bushman FD. Retroviral DNA integration -mechanism and consequences. Adv Genet. 2005;55:147–181. - PubMed
-
- Dyda F, et al. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. - PubMed
-
- Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell. 2005;121:1005–1016. - PubMed
-
- Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

