The effects of proteoglycan surface patterning on neuronal pathfinding
- PMID: 20119506
- PMCID: PMC2813059
- DOI: 10.1002/mawe.200700224
The effects of proteoglycan surface patterning on neuronal pathfinding
Abstract
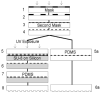
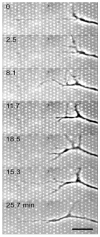
Protein micropatterning techniques are increasingly applied in cell choice assays to investigate fundamental biological phenomena that contribute to the host response to implanted biomaterials, and to explore the effects of protein stability and biological activity on cell behavior for in vitro cell studies. In the area of neuronal regeneration the protein micropatterning and cell choice assays are used to improve our understanding of the mechanisms directing nervous system during development and regenerative failure in the central nervous system (CNS) wound healing environment. In these cell assays, protein micropatterns need to be characterized for protein stability, bioactivity, and spatial distribution and then correlated with observed mammalian cell behavior using appropriate model system for CNS development and repair. This review provides the background on protein micropatterning for cell choice assays and describes some novel patterns that were developed to interrogate neuronal adaptation to inhibitory signals encountered in CNS injuries.
Figures




Similar articles
-
A fast and accessible methodology for micro-patterning cells on standard culture substrates using Parafilm™ inserts.PLoS One. 2011;6(6):e20909. doi: 10.1371/journal.pone.0020909. Epub 2011 Jun 7. PLoS One. 2011. PMID: 21687691 Free PMC article.
-
Zebrafish extracellular matrix improves neuronal viability and network formation in a 3-dimensional culture.Biomaterials. 2018 Jul;170:137-146. doi: 10.1016/j.biomaterials.2018.04.009. Epub 2018 Apr 6. Biomaterials. 2018. PMID: 29665503
-
Glycosaminoglycans' for brain health: Harnessing glycosaminoglycan based biomaterials for treating central nervous system diseases and in-vitro modeling.Biomaterials. 2024 Sep;309:122629. doi: 10.1016/j.biomaterials.2024.122629. Epub 2024 May 22. Biomaterials. 2024. PMID: 38797120 Review.
-
Natural recovery and regeneration of the central nervous system.Regen Med. 2022 Apr;17(4):233-244. doi: 10.2217/rme-2021-0084. Epub 2022 Feb 21. Regen Med. 2022. PMID: 35187979 Review.
-
Micropatterned composite membrane guides oriented cell growth and vascularization for accelerating wound healing.Regen Biomater. 2022 Dec 26;10:rbac108. doi: 10.1093/rb/rbac108. eCollection 2023. Regen Biomater. 2022. PMID: 36683746 Free PMC article.
Cited by
-
The influence of sub-micron inhibitory clusters on growth cone substratum attachments and CD44 expression.Biomaterials. 2008 Nov;29(31):4227-35. doi: 10.1016/j.biomaterials.2008.07.031. Epub 2008 Aug 9. Biomaterials. 2008. PMID: 18694596 Free PMC article.
-
Multiprotein microcontact printing with micrometer resolution.Langmuir. 2012 Jan 31;28(4):2238-43. doi: 10.1021/la2039202. Epub 2012 Jan 9. Langmuir. 2012. PMID: 22204564 Free PMC article.
-
Myelination and node of Ranvier formation on sensory neurons in a defined in vitro system.In Vitro Cell Dev Biol Anim. 2013 Sep;49(8):608-618. doi: 10.1007/s11626-013-9647-8. Epub 2013 Aug 16. In Vitro Cell Dev Biol Anim. 2013. PMID: 23949775 Free PMC article.
-
Sugar glues for broken neurons.Biomol Concepts. 2013 Jun;4(3):233-57. doi: 10.1515/bmc-2012-0042. Biomol Concepts. 2013. PMID: 25285176 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
