The deep intronic c.903+469T>C mutation in the MTRR gene creates an SF2/ASF binding exonic splicing enhancer, which leads to pseudoexon activation and causes the cblE type of homocystinuria
- PMID: 20120036
- PMCID: PMC3429857
- DOI: 10.1002/humu.21206
The deep intronic c.903+469T>C mutation in the MTRR gene creates an SF2/ASF binding exonic splicing enhancer, which leads to pseudoexon activation and causes the cblE type of homocystinuria
Abstract
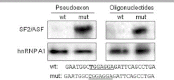
Deep intronic mutations are often ignored as possible causes of human diseases. A deep intronic mutation in the MTRR gene, c.903+469T>C, is the most frequent mutation causing the cblE type of homocystinuria. It is well known to be associated with pre-mRNA mis-splicing, resulting in pseudoexon inclusion; however, the pathological mechanism remains unknown. We used minigenes to demonstrate that this mutation is the direct cause of MTRR pseudoexon inclusion, and that the pseudoexon is normally not recognized due to a suboptimal 5' splice site. Within the pseudoexon we identified an exonic splicing enhancer (ESE), which is activated by the mutation. Cotransfection and siRNA experiments showed that pseudoexon inclusion depends on the cellular amounts of SF2/ASF and in vitro RNA-binding assays showed dramatically increased SF2/ASF binding to the mutant MTRR ESE. The mutant MTRR ESE sequence is identical to an ESE of the alternatively spliced MST1R proto-oncogene, which suggests that this ESE could be frequently involved in splicing regulation. Our study conclusively demonstrates that an intronic single nucleotide change is sufficient to cause pseudoexon activation via creation of a functional ESE, which binds a specific splicing factor. We suggest that this mechanism may cause genetic disease much more frequently than previously reported.
(c) 2010 Wiley-Liss, Inc.
Figures

Similar articles
-
Splice-shifting oligonucleotide (SSO) mediated blocking of an exonic splicing enhancer (ESE) created by the prevalent c.903+469T>C MTRR mutation corrects splicing and restores enzyme activity in patient cells.Nucleic Acids Res. 2015 May 19;43(9):4627-39. doi: 10.1093/nar/gkv275. Epub 2015 Apr 15. Nucleic Acids Res. 2015. PMID: 25878036 Free PMC article.
-
A deep intronic mutation in FGB creates a consensus exonic splicing enhancer motif that results in afibrinogenemia caused by aberrant mRNA splicing, which can be corrected in vitro with antisense oligonucleotide treatment.Hum Mutat. 2009 Feb;30(2):221-7. doi: 10.1002/humu.20839. Hum Mutat. 2009. PMID: 18853456
-
Deeper understanding of unclassified intronic variants and ESEs.Hum Mutat. 2010 Apr;31(4):V. doi: 10.1002/humu.21247. Hum Mutat. 2010. PMID: 20358580 No abstract available.
-
Pseudoexon activation in disease by non-splice site deep intronic sequence variation - wild type pseudoexons constitute high-risk sites in the human genome.Hum Mutat. 2022 Feb;43(2):103-127. doi: 10.1002/humu.24306. Epub 2021 Dec 5. Hum Mutat. 2022. PMID: 34837434 Review.
-
Splicing mutations in inherited retinal diseases.Prog Retin Eye Res. 2021 Jan;80:100874. doi: 10.1016/j.preteyeres.2020.100874. Epub 2020 Jun 15. Prog Retin Eye Res. 2021. PMID: 32553897 Review.
Cited by
-
The Asthma-associated PER1-like domain-containing protein 1 (PERLD1) Haplotype Influences Soluble Glycosylphosphatidylinositol Anchor Protein (sGPI-AP) Levels in Serum and Immune Cell Proliferation.Sci Rep. 2020 Jan 20;10(1):715. doi: 10.1038/s41598-020-57592-9. Sci Rep. 2020. PMID: 31959860 Free PMC article.
-
Genome-wide association study identifies phospholipase C zeta 1 (PLCz1) as a stallion fertility locus in Hanoverian warmblood horses.PLoS One. 2014 Oct 29;9(10):e109675. doi: 10.1371/journal.pone.0109675. eCollection 2014. PLoS One. 2014. PMID: 25354211 Free PMC article.
-
Antisense Oligonucleotide Screening to Optimize the Rescue of the Splicing Defect Caused by the Recurrent Deep-Intronic ABCA4 Variant c.4539+2001G>A in Stargardt Disease.Genes (Basel). 2019 Jun 14;10(6):452. doi: 10.3390/genes10060452. Genes (Basel). 2019. PMID: 31197102 Free PMC article.
-
Novel GUCA1A mutations suggesting possible mechanisms of pathogenesis in cone, cone-rod, and macular dystrophy patients.Biomed Res Int. 2013;2013:517570. doi: 10.1155/2013/517570. Epub 2013 Aug 14. Biomed Res Int. 2013. PMID: 24024198 Free PMC article.
-
Deep intronic mutations and human disease.Hum Genet. 2017 Sep;136(9):1093-1111. doi: 10.1007/s00439-017-1809-4. Epub 2017 May 12. Hum Genet. 2017. PMID: 28497172 Review.
References
-
- Akker SA, Misra S, Aslam S, Morgan EL, Smith PJ, Khoo B, Chew SL. Pre-spliceosomal binding of U1 small nuclear ribonucleoprotein (RNP) and heterogenous nuclear RNP E1 is associated with suppression of a growth hormone receptor pseudoexon. Mol Endocrinol. 2007;21:2529–2540. - PubMed
-
- Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. - PubMed
-
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

