Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening
- PMID: 20121196
- PMCID: PMC2842983
- DOI: 10.1021/jm901386e
Discovery of novel fibroblast growth factor receptor 1 kinase inhibitors by structure-based virtual screening
Abstract
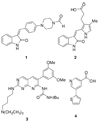
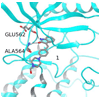
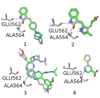
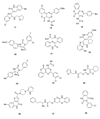
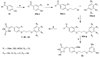
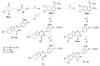
Fibroblast growth factors (FGFs) play important roles in embryonic development, angiogenesis, wound healing, and cell proliferation and differentiation. In search of inhibitors of FGFR1 kinase, 2.2 million compounds were docked into the ATP binding site of the protein. A co-crystal structure, which shows two alternative conformations for the nucleotide binding loop, is reported. Docking was performed on both conformations and, ultimately, 23 diverse compounds were purchased and assayed. Following hit validation, two compounds 10 and 16, a benzylidene derivative of pseudothiohydantoin and a thienopyrimidinone derivative, respectively, were discovered that inhibit FGFR1 kinase with IC(50) values of 23 and 50 microM. Initial optimization of 16 led to the more unsaturated 40, which has significantly enhanced potency, 1.9 microM. The core structures represent new structural motifs for FGFR1 kinase inhibitors. The study also illustrates complexities associated with the choice of protein structures for docking, possible use of multiple kinase structures to seek selectivity, and hit identification.
Figures


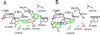
Similar articles
-
A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.J Biol Chem. 2011 Jun 10;286(23):20677-87. doi: 10.1074/jbc.M110.213736. Epub 2011 Mar 24. J Biol Chem. 2011. PMID: 21454610 Free PMC article.
-
Design, synthesis and biological evaluation of novel 1H-1,2,4-triazole, benzothiazole and indazole-based derivatives as potent FGFR1 inhibitors viafragment-based virtual screening.J Enzyme Inhib Med Chem. 2020 Dec;35(1):72-84. doi: 10.1080/14756366.2019.1673745. J Enzyme Inhib Med Chem. 2020. PMID: 31682465 Free PMC article.
-
Design, Synthesis, and Biological Evaluation of C-2 Substituted 3Hthieno[ 2,3-d]pyrimidin-4-one Derivatives as Novel FGFR1 Inhibitors.Med Chem. 2017;13(8):753-760. doi: 10.2174/1573406413666170623084525. Med Chem. 2017. PMID: 28641527
-
Improved lead-finding for kinase targets using high-throughput docking.Curr Opin Drug Discov Devel. 2006 May;9(3):339-47. Curr Opin Drug Discov Devel. 2006. PMID: 16729730 Review.
-
Discovery and optimization of novel pyrazole-benzimidazole CPL304110, as a potent and selective inhibitor of fibroblast growth factor receptors FGFR (1-3).Eur J Med Chem. 2021 Jan 15;210:112990. doi: 10.1016/j.ejmech.2020.112990. Epub 2020 Nov 7. Eur J Med Chem. 2021. PMID: 33199155 Review.
Cited by
-
Alkoxyalkylation of Electron-Rich Aromatic Compounds.Int J Mol Sci. 2024 Jun 26;25(13):6966. doi: 10.3390/ijms25136966. Int J Mol Sci. 2024. PMID: 39000077 Free PMC article. Review.
-
Absolute binding free energy calculations improve enrichment of actives in virtual compound screening.Sci Rep. 2022 Aug 10;12(1):13640. doi: 10.1038/s41598-022-17480-w. Sci Rep. 2022. PMID: 35948614 Free PMC article.
-
Identification and validation of novel PERK inhibitors.J Chem Inf Model. 2014 May 27;54(5):1467-75. doi: 10.1021/ci500114r. Epub 2014 May 5. J Chem Inf Model. 2014. PMID: 24745945 Free PMC article.
-
Aryl Extensions of Thienopyrimidinones as Fibroblast Growth Factor Receptor 1 Kinase Inhibitors.Tetrahedron Lett. 2011 Apr 27;52(17):2228-2231. doi: 10.1016/j.tetlet.2010.12.081. Tetrahedron Lett. 2011. PMID: 21516197 Free PMC article.
-
A novel mode of protein kinase inhibition exploiting hydrophobic motifs of autoinhibited kinases: discovery of ATP-independent inhibitors of fibroblast growth factor receptor.J Biol Chem. 2011 Jun 10;286(23):20677-87. doi: 10.1074/jbc.M110.213736. Epub 2011 Mar 24. J Biol Chem. 2011. PMID: 21454610 Free PMC article.
References
-
- Bishop JM. The molecular genetics of cancer. Science. 1987;235:305–311. - PubMed
-
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990;61:203–212. - PubMed
-
- Aaronson AS. Growth factors and cancer. Science. 1991;254:1146–1153. - PubMed
-
- Jaye M, Schlessinger J, Dionne C. Fibroblast growth factor receptor tyrosine kinases: molecular analysis and signal transduction. Biochim. Biophys. Acta. 1992;1135:185–199. - PubMed
-
- Basilico C, Moscatelli D. The Fgf Family of Growth Factors and Oncogenes. Adv. Cancer Res. 1992;59:115–165. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

