Quiescin sulfhydryl oxidase from Trypanosoma brucei: catalytic activity and mechanism of a QSOX family member with a single thioredoxin domain
- PMID: 20121244
- PMCID: PMC2831140
- DOI: 10.1021/bi902222s
Quiescin sulfhydryl oxidase from Trypanosoma brucei: catalytic activity and mechanism of a QSOX family member with a single thioredoxin domain
Abstract
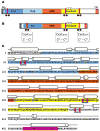
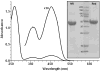
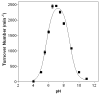
Quiescin sulfhydryl oxidase (QSOX) flavoenzymes catalyze the direct, facile, insertion of disulfide bonds into reduced unfolded proteins with the reduction of oxygen to hydrogen peroxide. To date, only QSOXs from vertebrates have been characterized enzymatically. These metazoan sulfhydryl oxidases have four recognizable domains: a redox-active thioredoxin (Trx) domain containing the first of three CxxC motifs (C(I)-C(II)), a second Trx domain with no obvious redox-active disulfide, a helix-rich domain, and then an Erv/ALR domain. This last domain contains the FAD moiety, a proximal C(III)-C(IV) disulfide, and a third CxxC of unknown function (C(V)-C(VI)). Plant and protist QSOXs lack the second Trx domain but otherwise appear to contain the same complement of redox centers. This work presents the first characterization of a single-Trx QSOX. Trypanosoma brucei QSOX was expressed in Escherichia coli using a synthetic gene and found to be a stable, monomeric, FAD-containing protein. Although evidently lacking an entire domain, TbQSOX shows catalytic activity and substrate specificity similar to the vertebrate QSOXs examined previously. Unfolded reduced proteins are more than 200-fold more effective substrates on a per thiol basis than glutathione and some 10-fold better than the parasite bisglutathione analogue, trypanothione. These data are consistent with a role for the protist QSOX in oxidative protein folding. Site-directed mutagenesis of each of the six cysteine residues (to serines) shows that the CxxC motif in the single-Trx domain is crucial for efficient catalysis of the oxidation of both reduced RNase and the model substrate dithiothreitol. As expected, the proximal disulfide C(III)-C(IV), which interacts with the flavin, is catalytically crucial. However, as observed with human QSOX1, the third CxxC motif shows no obvious catalytic role during the in vitro oxidation of reduced RNase or dithiothreitol. Pre-steady-state kinetics demonstrates that turnover in TbQSOX is limited by an internal redox step leading to 2-electron reduction of the FAD cofactor. In sum, the single-Trx domain QSOX studied here shows a striking similarity in enzymatic behavior to its double-Trx metazoan counterparts.
Figures
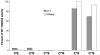
Similar articles
-
Going through the barrier: coupled disulfide exchange reactions promote efficient catalysis in quiescin sulfhydryl oxidase.J Biol Chem. 2014 Feb 21;289(8):5274-84. doi: 10.1074/jbc.M113.536219. Epub 2013 Dec 30. J Biol Chem. 2014. PMID: 24379406 Free PMC article.
-
Human quiescin-sulfhydryl oxidase, QSOX1: probing internal redox steps by mutagenesis.Biochemistry. 2008 Apr 29;47(17):4955-63. doi: 10.1021/bi702522q. Epub 2008 Apr 5. Biochemistry. 2008. PMID: 18393449 Free PMC article.
-
Inter-domain redox communication in flavoenzymes of the quiescin/sulfhydryl oxidase family: role of a thioredoxin domain in disulfide bond formation.Biochemistry. 2003 Apr 22;42(15):4560-8. doi: 10.1021/bi030003z. Biochemistry. 2003. PMID: 12693953
-
Oxidative protein folding and the Quiescin-sulfhydryl oxidase family of flavoproteins.Antioxid Redox Signal. 2010 Oct;13(8):1217-30. doi: 10.1089/ars.2010.3098. Antioxid Redox Signal. 2010. PMID: 20136510 Free PMC article. Review.
-
Generating disulfides with the Quiescin-sulfhydryl oxidases.Biochim Biophys Acta. 2008 Apr;1783(4):567-77. doi: 10.1016/j.bbamcr.2007.10.002. Epub 2007 Oct 12. Biochim Biophys Acta. 2008. PMID: 17980160 Free PMC article. Review.
Cited by
-
Elevated transcription of the gene QSOX1 encoding quiescin Q6 sulfhydryl oxidase 1 in breast cancer.PLoS One. 2013;8(2):e57327. doi: 10.1371/journal.pone.0057327. Epub 2013 Feb 27. PLoS One. 2013. PMID: 23460839 Free PMC article.
-
Ebselen inhibits QSOX1 enzymatic activity and suppresses invasion of pancreatic and renal cancer cell lines.Oncotarget. 2015 Jul 30;6(21):18418-28. doi: 10.18632/oncotarget.4099. Oncotarget. 2015. PMID: 26158899 Free PMC article.
-
Protein substrate discrimination in the quiescin sulfhydryl oxidase (QSOX) family.Biochemistry. 2012 May 22;51(20):4226-35. doi: 10.1021/bi300394w. Epub 2012 May 14. Biochemistry. 2012. PMID: 22582951 Free PMC article.
-
The emerging role of QSOX1 in cancer.Antioxid Redox Signal. 2014 Jul 20;21(3):485-96. doi: 10.1089/ars.2013.5572. Epub 2014 Feb 19. Antioxid Redox Signal. 2014. PMID: 24359107 Free PMC article. Review.
-
Slow domain reconfiguration causes power-law kinetics in a two-state enzyme.Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):513-518. doi: 10.1073/pnas.1714401115. Epub 2018 Jan 3. Proc Natl Acad Sci U S A. 2018. PMID: 29298911 Free PMC article.
References
-
- Thorpe C, Hoober K, Raje S, Glynn N, Burnside J, Turi G, Coppock D. Sulfhydryl oxidases: emerging catalysts of protein disulfide bond formation in eukaryotes. Arch Biochem Biophys. 2002;405:1–12. - PubMed
-
- Thorpe C, Coppock DL. Generating disulfides in multicellular organisms: Emerging roles for a new flavoprotein family. J Biol Chem. 2007;282:13929–13933. - PubMed
-
- Coppock DL, Thorpe C. Multidomain flavin-dependent sulfhydryl oxidases. Antioxid Redox Signal. 2006;8:300–311. - PubMed
-
- Ostrowski MC, Kistler WS. Properties of a flavoprotein sulfhydryl oxidase from rat seminal vesicle secretion. Biochemistry. 1980;19:2639–2645. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

