Comparative evolutionary analysis of protein complexes in E. coli and yeast
- PMID: 20122144
- PMCID: PMC2837643
- DOI: 10.1186/1471-2164-11-79
Comparative evolutionary analysis of protein complexes in E. coli and yeast
Abstract
Background: Proteins do not act in isolation; they frequently act together in protein complexes to carry out concerted cellular functions. The evolution of complexes is poorly understood, especially in organisms other than yeast, where little experimental data has been available.
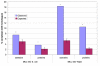
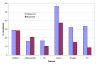
Results: We generated accurate, high coverage datasets of protein complexes for E. coli and yeast in order to study differences in the evolution of complexes between these two species. We show that substantial differences exist in how complexes have evolved between these organisms. A previously proposed model of complex evolution identified complexes with cores of interacting homologues. We support findings of the relative importance of this mode of evolution in yeast, but find that it is much less common in E. coli. Additionally it is shown that those homologues which do cluster in complexes are involved in eukaryote-specific functions. Furthermore we identify correlated pairs of non-homologous domains which occur in multiple protein complexes. These were identified in both yeast and E. coli and we present evidence that these too may represent complex cores in yeast but not those of E. coli.
Conclusions: Our results suggest that there are differences in the way protein complexes have evolved in E. coli and yeast. Whereas some yeast complexes have evolved by recruiting paralogues, this is not apparent in E. coli. Furthermore, such complexes are involved in eukaryotic-specific functions. This implies that the increase in gene family sizes seen in eukaryotes in part reflects multiple family members being used within complexes. However, in general, in both E. coli and yeast, homologous domains are used in different complexes.
Figures
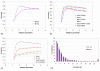
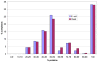
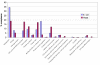
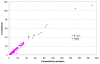
Similar articles
-
Evolutionary rate heterogeneity of core and attachment proteins in yeast protein complexes.Genome Biol Evol. 2013;5(7):1366-75. doi: 10.1093/gbe/evt096. Genome Biol Evol. 2013. PMID: 23814130 Free PMC article.
-
Detection of overlapping protein complexes in gene expression, phenotype and pathways of Saccharomyces cerevisiae using Prorank based Fuzzy algorithm.Gene. 2016 Apr 15;580(2):144-158. doi: 10.1016/j.gene.2016.01.016. Epub 2016 Jan 22. Gene. 2016. PMID: 26809099
-
Predicting protein complexes from weighted protein-protein interaction graphs with a novel unsupervised methodology: Evolutionary enhanced Markov clustering.Artif Intell Med. 2015 Mar;63(3):181-9. doi: 10.1016/j.artmed.2014.12.012. Epub 2015 Feb 18. Artif Intell Med. 2015. PMID: 25765008
-
A survey of computational methods for protein complex prediction from protein interaction networks.J Bioinform Comput Biol. 2013 Apr;11(2):1230002. doi: 10.1142/S021972001230002X. Epub 2012 Nov 7. J Bioinform Comput Biol. 2013. PMID: 23600810 Review.
-
Insights into molecular evolution from yeast genomics.Yeast. 2014 Jul;31(7):233-41. doi: 10.1002/yea.3018. Epub 2014 May 26. Yeast. 2014. PMID: 24760744 Review.
Cited by
-
Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states.Proc Natl Acad Sci U S A. 2010 Nov 23;107(47):20352-7. doi: 10.1073/pnas.1012999107. Epub 2010 Nov 3. Proc Natl Acad Sci U S A. 2010. PMID: 21048085 Free PMC article.
-
The evolution of multimeric protein assemblages.Mol Biol Evol. 2012 May;29(5):1353-66. doi: 10.1093/molbev/msr300. Epub 2011 Dec 5. Mol Biol Evol. 2012. PMID: 22144639 Free PMC article.
-
Diverging co-translational protein complex assembly pathways are governed by interface energy distribution.Nat Commun. 2024 Mar 25;15(1):2638. doi: 10.1038/s41467-024-46881-w. Nat Commun. 2024. PMID: 38528060 Free PMC article.
-
Diversity in protein domain superfamilies.Curr Opin Genet Dev. 2015 Dec;35:40-9. doi: 10.1016/j.gde.2015.09.005. Epub 2015 Nov 3. Curr Opin Genet Dev. 2015. PMID: 26451979 Free PMC article. Review.
-
Functional site plasticity in domain superfamilies.Biochim Biophys Acta. 2013 May;1834(5):874-89. doi: 10.1016/j.bbapap.2013.02.042. Epub 2013 Mar 14. Biochim Biophys Acta. 2013. PMID: 23499848 Free PMC article.
References
-
- Ishihama A. Functional modulation of Escherichia coli RNA polymerase. AnnuRevMicrobiol. 2000;54:499–518. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

