The mutation T477A in HIV-1 reverse transcriptase (RT) restores normal proteolytic processing of RT in virus with Gag-Pol mutated in the p51-RNH cleavage site
- PMID: 20122159
- PMCID: PMC2831009
- DOI: 10.1186/1742-4690-7-6
The mutation T477A in HIV-1 reverse transcriptase (RT) restores normal proteolytic processing of RT in virus with Gag-Pol mutated in the p51-RNH cleavage site
Abstract
Background: The p51 subunit of the HIV-1 reverse transcriptase (RT) p66/p51 heterodimer arises from proteolytic cleavage of the RT p66 subunit C-terminal ribonuclease H (RNH) domain during virus maturation. Our previous work showed that mutations in the RT p51 downward arrowRNH cleavage site resulted in virus with defects in proteolytic processing of RT and significantly attenuated infectivity. In some cases, virus fitness was restored after repeated passage of mutant viruses, due to reversion of the mutated sequences to wild-type. However, in one case, the recovered virus retained the mutated p51 downward arrowRNH cleavage site but also developed an additional mutation, T477A, distal to the cleavage site. In this study we have characterized in detail the impact of the T477A mutation on intravirion processing of RT.
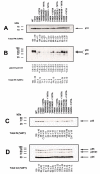
Results: While the T477A mutation arose during serial passage only with the F440V mutant background, introduction of this substitution into a variety of RT p51 downward arrowRNH cleavage site lethal mutant backgrounds was able to restore substantial infectivity and normal RT processing to these mutants. T477A had no phenotypic effect on wild-type HIV-1. We also evaluated the impact of T477A on the kinetics of intravirion Gag-Pol polyprotein processing of p51 downward arrowRNH cleavage site mutants using the protease inhibitor ritonavir. Early processing intermediates accumulated in p51 downward arrowRNH cleavage site mutant viruses, whereas introduction of T477A promoted the completion of processing and formation of the fully processed RT p66/p51 heterodimer.
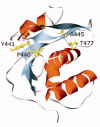
Conclusions: This work highlights the extraordinary plasticity of HIV-1 in adapting to seemingly lethal mutations that prevent RT heterodimer formation during virion polyprotein maturation. The ability of T477A to restore RT heterodimer formation and thus intravirion stability of the enzyme may arise from increased conformation flexibility in the RT p51 downward arrowRNH cleavage site region, due to loss of a hydrogen bond associated with the normal threonine residue, thereby enabling proteolytic cleavage near the normal RT p51 downward arrowRNH cleavage site.
Figures


References
-
- Chattopadhyay D, Evans DB, Deibel MR Jr. Purification and characterization of heterodimeric human immunodeficiency virus type 1 (HIV-1) reverse transcriptase produced by in vitro processing of p66 with recombinant HIV-1 protease. J Biol Chem. 1992;267(20):14227–32. - PubMed
-
- Fan N, Rank KB, Leone JW. The differential processing of homodimers of reverse transcriptases from human immunodeficiency viruses type 1 and 2 is a consequence of the distinct specificities of the viral proteases. J Biol Chem. 1995;270(22):13573–9. - PubMed
-
- Hostomska Z, Matthews DA, Davies JF, Nodes BR, Hostomsky Z. Proteolytic release and crystallization of the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1991;266(22):14697–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

