Predicting the protein-protein interactions using primary structures with predicted protein surface
- PMID: 20122202
- PMCID: PMC3009501
- DOI: 10.1186/1471-2105-11-S1-S3
Predicting the protein-protein interactions using primary structures with predicted protein surface
Abstract
Background: Many biological functions involve various protein-protein interactions (PPIs). Elucidating such interactions is crucial for understanding general principles of cellular systems. Previous studies have shown the potential of predicting PPIs based on only sequence information. Compared to approaches that require other auxiliary information, these sequence-based approaches can be applied to a broader range of applications.
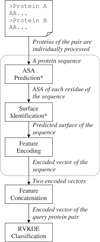
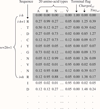
Results: This study presents a novel sequence-based method based on the assumption that protein-protein interactions are more related to amino acids at the surface than those at the core. The present method considers surface information and maintains the advantage of relying on only sequence data by including an accessible surface area (ASA) predictor recently proposed by the authors. This study also reports the experiments conducted to evaluate a) the performance of PPI prediction achieved by including the predicted surface and b) the quality of the predicted surface in comparison with the surface obtained from structures. The experimental results show that surface information helps to predict interacting protein pairs. Furthermore, the prediction performance achieved by using the surface estimated with the ASA predictor is close to that using the surface obtained from protein structures.
Conclusion: This work presents a sequence-based method that takes into account surface information for predicting PPIs. The proposed procedure of surface identification improves the prediction performance with an F-measure of 5.1%. The extracted surfaces are also valuable in other biomedical applications that require similar information.
Figures


Similar articles
-
Predicting protein-protein interactions in unbalanced data using the primary structure of proteins.BMC Bioinformatics. 2010 Apr 2;11:167. doi: 10.1186/1471-2105-11-167. BMC Bioinformatics. 2010. PMID: 20361868 Free PMC article.
-
A discriminative approach for identifying domain-domain interactions from protein-protein interactions.Proteins. 2010 Apr;78(5):1243-53. doi: 10.1002/prot.22643. Proteins. 2010. PMID: 20027642
-
A Cascade Random Forests Algorithm for Predicting Protein-Protein Interaction Sites.IEEE Trans Nanobioscience. 2015 Oct;14(7):746-60. doi: 10.1109/TNB.2015.2475359. Epub 2015 Sep 28. IEEE Trans Nanobioscience. 2015. PMID: 26441427
-
Predicting protein function from sequence and structural data.Curr Opin Struct Biol. 2005 Jun;15(3):275-84. doi: 10.1016/j.sbi.2005.04.003. Curr Opin Struct Biol. 2005. PMID: 15963890 Review.
-
A survey on computational models for predicting protein-protein interactions.Brief Bioinform. 2021 Sep 2;22(5):bbab036. doi: 10.1093/bib/bbab036. Brief Bioinform. 2021. PMID: 33693513 Review.
Cited by
-
Combining phylogenetic profiling-based and machine learning-based techniques to predict functional related proteins.PLoS One. 2013 Sep 19;8(9):e75940. doi: 10.1371/journal.pone.0075940. eCollection 2013. PLoS One. 2013. PMID: 24069454 Free PMC article.
-
SPRINT: ultrafast protein-protein interaction prediction of the entire human interactome.BMC Bioinformatics. 2017 Nov 15;18(1):485. doi: 10.1186/s12859-017-1871-x. BMC Bioinformatics. 2017. PMID: 29141584 Free PMC article.
-
Fundamentals of protein interaction network mapping.Mol Syst Biol. 2015 Dec 17;11(12):848. doi: 10.15252/msb.20156351. Mol Syst Biol. 2015. PMID: 26681426 Free PMC article. Review.
-
On protocols and measures for the validation of supervised methods for the inference of biological networks.Front Genet. 2013 Dec 3;4:262. doi: 10.3389/fgene.2013.00262. Front Genet. 2013. PMID: 24348517 Free PMC article. Review.
-
PPIcons: identification of protein-protein interaction sites in selected organisms.J Mol Model. 2013 Sep;19(9):4059-70. doi: 10.1007/s00894-013-1886-9. Epub 2013 Jun 2. J Mol Model. 2013. PMID: 23729008 Free PMC article.
References
-
- Tong AHY, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S. et al.A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science. 2002;295(5553):321–324. doi: 10.1126/science.1064987. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

