Computational prediction of type III secreted proteins from gram-negative bacteria
- PMID: 20122221
- PMCID: PMC3009519
- DOI: 10.1186/1471-2105-11-S1-S47
Computational prediction of type III secreted proteins from gram-negative bacteria
Abstract
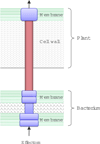
Background: Type III secretion system (T3SS) is a specialized protein delivery system in gram-negative bacteria that injects proteins (called effectors) directly into the eukaryotic host cytosol and facilitates bacterial infection. For many plant and animal pathogens, T3SS is indispensable for disease development. Recently, T3SS has also been found in rhizobia and plays a crucial role in the nodulation process. Although a great deal of efforts have been done to understand type III secretion, the precise mechanism underlying the secretion and translocation process has not been fully understood. In particular, defined secretion and translocation signals enabling the secretion have not been identified from the type III secreted effectors (T3SEs), which makes the identification of these important virulence factors notoriously challenging. The availability of a large number of sequenced genomes for plant and animal-associated bacteria demands the development of efficient and effective prediction methods for the identification of T3SEs using bioinformatics approaches.
Results: We have developed a machine learning method based on the N-terminal amino acid sequences to predict novel type III effectors in the plant pathogen Pseudomonas syringae and the microsymbiont rhizobia. The extracted features used in the learning model (or classifier) include amino acid composition, secondary structure and solvent accessibility information. The method achieved a precision of over 90% on P. syringae in a cross validation study. In combination with a promoter screen for the type III specific promoters, this classifier trained on the P. syringae data was applied to predict novel T3SEs from the genomic sequences of four rhizobial strains. This application resulted in 57 candidate type III secreted proteins, 17 of which are confirmed effectors.
Conclusion: Our experimental results demonstrate that the machine learning method based on N-terminal amino acid sequences combined with a promoter screen could prove to be a very effective computational approach for predicting novel type III effectors in gram-negative bacteria. Our method and data are available to the public upon request.
Figures
Similar articles
-
Identification of novel type III effectors using latent Dirichlet allocation.Comput Math Methods Med. 2012;2012:696190. doi: 10.1155/2012/696190. Epub 2012 Sep 2. Comput Math Methods Med. 2012. PMID: 22997537 Free PMC article.
-
A new feature selection method for computational prediction of type III secreted effectors.Int J Data Min Bioinform. 2014;10(4):440-54. doi: 10.1504/ijdmb.2014.064894. Int J Data Min Bioinform. 2014. PMID: 25946888
-
Functional and computational analysis of amino acid patterns predictive of type III secretion system substrates in Pseudomonas syringae.PLoS One. 2012;7(4):e36038. doi: 10.1371/journal.pone.0036038. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558318 Free PMC article.
-
Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains.Mol Plant Microbe Interact. 2006 Nov;19(11):1151-8. doi: 10.1094/MPMI-19-1151. Mol Plant Microbe Interact. 2006. PMID: 17073298 Review.
-
Computational prediction of type III and IV secreted effectors in gram-negative bacteria.Infect Immun. 2011 Jan;79(1):23-32. doi: 10.1128/IAI.00537-10. Epub 2010 Oct 25. Infect Immun. 2011. PMID: 20974833 Free PMC article. Review.
Cited by
-
SrfJ, a Salmonella type III secretion system effector regulated by PhoP, RcsB, and IolR.J Bacteriol. 2012 Aug;194(16):4226-36. doi: 10.1128/JB.00173-12. Epub 2012 Jun 1. J Bacteriol. 2012. PMID: 22661691 Free PMC article.
-
Analysis of the expression, secretion and translocation of the Salmonella enterica type III secretion system effector SteA.PLoS One. 2011;6(10):e26930. doi: 10.1371/journal.pone.0026930. Epub 2011 Oct 27. PLoS One. 2011. PMID: 22046414 Free PMC article.
-
Effective identification of Gram-negative bacterial type III secreted effectors using position-specific residue conservation profiles.PLoS One. 2013 Dec 31;8(12):e84439. doi: 10.1371/journal.pone.0084439. eCollection 2013. PLoS One. 2013. PMID: 24391954 Free PMC article.
-
DeepT3_4: A Hybrid Deep Neural Network Model for the Distinction Between Bacterial Type III and IV Secreted Effectors.Front Microbiol. 2021 Jan 21;12:605782. doi: 10.3389/fmicb.2021.605782. eCollection 2021. Front Microbiol. 2021. PMID: 33552038 Free PMC article.
-
DeepT3 2.0: improving type III secreted effector predictions by an integrative deep learning framework.NAR Genom Bioinform. 2021 Oct 4;3(4):lqab086. doi: 10.1093/nargab/lqab086. eCollection 2021 Dec. NAR Genom Bioinform. 2021. PMID: 34617013 Free PMC article.
References
-
- He SY, Nomura K, Whittam TS. Type III protein secretion mechanism in mammalian and plant pathogens. BBA-Molecular Cell Research. 2004;1694(1-3):181–206. - PubMed