Comparison of PGH2 binding site in prostaglandin synthases
- PMID: 20122226
- PMCID: PMC3009524
- DOI: 10.1186/1471-2105-11-S1-S51
Comparison of PGH2 binding site in prostaglandin synthases
Abstract
Background: Prostaglandin H2 (PGH2) is a common precursor for the synthesis of five different Prostanoids via specific Prostanoid Synthases. The binding of this substrate with these Synthases is not properly understood. Moreover, currently no crystal structure of complexes bound with PGH2 has been reported. Hence, understanding the interactions of PGH2 and characterizing its binding sites in these synthases is crucial for developing novel therapeutics based on these proteins as targets.
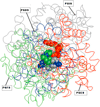
Results: Shape and physico-chemical properties of the PGH2 binding sites of the four prostanoid synthases were analyzed and compared in order to understand the molecular basis of the specificity. This study provides models with predicted pockets for the binding of PGH2 with PGD, PGE, PGF and PGI Synthases. The results closely match with available experimental data. The comparison showed seven physico-chemical features that are common to the four PGH2 binding sites. However this common pattern is not statistically unique and is not specific enough to distinguish between proteins that can or cannot bind PGH2. A large scale search in ASTRAL data bank, a non redundant Protein Data Bank, for a similar pattern showed the uniqueness of each of the PGH2 binding site in these Synthases.
Conclusion: The binding pockets in PGDS, PGES, PGFS and PGIS are unique and do not share significant commonality which can be characterized as a PGH2 binding site. Local comparison of these protein structures highlights a case of convergent evolution in analogous functional sites.
Figures

Similar articles
-
PGH2 degradation pathway catalyzed by GSH-heme complex bound microsomal prostaglandin E2 synthase type 2: the first example of a dual-function enzyme.Biochemistry. 2007 Jul 17;46(28):8414-24. doi: 10.1021/bi700605m. Epub 2007 Jun 22. Biochemistry. 2007. PMID: 17585783
-
Structural and functional characterization of human microsomal prostaglandin E synthase-1 by computational modeling and site-directed mutagenesis.Bioorg Med Chem. 2006 May 15;14(10):3553-62. doi: 10.1016/j.bmc.2006.01.010. Epub 2006 Jan 24. Bioorg Med Chem. 2006. PMID: 16439136
-
Rate of vasoconstrictor prostanoids released by endothelial cells depends on cyclooxygenase-2 expression and prostaglandin I synthase activity.Circ Res. 1998 Aug 24;83(4):353-65. doi: 10.1161/01.res.83.4.353. Circ Res. 1998. PMID: 9721692
-
[Current topics in the regulation of prostanoids--3. Prostanoid synthases and prostanoid receptors].Masui. 1999 Jan;48(1):42-7. Masui. 1999. PMID: 10036888 Review. Japanese.
-
The structures of prostaglandin endoperoxide H synthases-1 and -2.Prostaglandins Other Lipid Mediat. 2002 Aug;68-69:129-52. doi: 10.1016/s0090-6980(02)00026-6. Prostaglandins Other Lipid Mediat. 2002. PMID: 12432914 Review.
Cited by
-
Exploration of binding site pattern in arachidonic acid metabolizing enzymes, Cyclooxygenases and Lipoxygenases.BMC Res Notes. 2015 Apr 16;8:152. doi: 10.1186/s13104-015-1101-4. BMC Res Notes. 2015. PMID: 25886468 Free PMC article.
-
Molecular docking studies and biological evaluation of isoxazole-carboxamide derivatives as COX inhibitors and antimicrobial agents.3 Biotech. 2022 Dec;12(12):342. doi: 10.1007/s13205-022-03408-8. Epub 2022 Nov 5. 3 Biotech. 2022. PMID: 36345437 Free PMC article.
-
Design, synthesis, molecular docking studies and biological evaluation of thiazole carboxamide derivatives as COX inhibitors.BMC Chem. 2023 Mar 6;17(1):11. doi: 10.1186/s13065-023-00924-3. BMC Chem. 2023. PMID: 36879343 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

