Active machine learning for transmembrane helix prediction
- PMID: 20122233
- PMCID: PMC3009531
- DOI: 10.1186/1471-2105-11-S1-S58
Active machine learning for transmembrane helix prediction
Abstract
Background: About 30% of genes code for membrane proteins, which are involved in a wide variety of crucial biological functions. Despite their importance, experimentally determined structures correspond to only about 1.7% of protein structures deposited in the Protein Data Bank due to the difficulty in crystallizing membrane proteins. Algorithms that can identify proteins whose high-resolution structure can aid in predicting the structure of many previously unresolved proteins are therefore of potentially high value. Active machine learning is a supervised machine learning approach which is suitable for this domain where there are a large number of sequences but only very few have known corresponding structures. In essence, active learning seeks to identify proteins whose structure, if revealed experimentally, is maximally predictive of others.
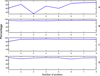
Results: An active learning approach is presented for selection of a minimal set of proteins whose structures can aid in the determination of transmembrane helices for the remaining proteins. TMpro, an algorithm for high accuracy TM helix prediction we previously developed, is coupled with active learning. We show that with a well-designed selection procedure, high accuracy can be achieved with only few proteins. TMpro, trained with a single protein achieved an F-score of 94% on benchmark evaluation and 91% on MPtopo dataset, which correspond to the state-of-the-art accuracies on TM helix prediction that are achieved usually by training with over 100 training proteins.
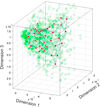
Conclusion: Active learning is suitable for bioinformatics applications, where manually characterized data are not a comprehensive representation of all possible data, and in fact can be a very sparse subset thereof. It aids in selection of data instances which when characterized experimentally can improve the accuracy of computational characterization of remaining raw data. The results presented here also demonstrate that the feature extraction method of TMpro is well designed, achieving a very good separation between TM and non TM segments.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

