NeMo: Network Module identification in Cytoscape
- PMID: 20122237
- PMCID: PMC3009535
- DOI: 10.1186/1471-2105-11-S1-S61
NeMo: Network Module identification in Cytoscape
Abstract
Background: As the size of the known human interactome grows, biologists increasingly rely on computational tools to identify patterns that represent protein complexes and pathways. Previous studies have shown that densely connected network components frequently correspond to community structure and functionally related modules. In this work, we present a novel method to identify densely connected and bipartite network modules based on a log odds score for shared neighbours.
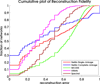
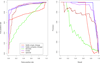
Results: To evaluate the performance of our method (NeMo), we compare it to other widely used tools for community detection including kMetis, MCODE, and spectral clustering. We test these methods on a collection of synthetically constructed networks and the set of MIPS human complexes. We apply our method to the CXC chemokine pathway and find a high scoring functional module of 12 disconnected phospholipase isoforms.
Conclusion: We present a novel method that combines a unique neighbour-sharing score with hierarchical agglomerative clustering to identify diverse network communities. The approach is unique in that we identify both dense network and dense bipartite network structures in a single approach. Our results suggest that the performance of NeMo is better than or competitive with leading approaches on both real and synthetic datasets. We minimize model complexity and generalization error in the Bayesian spirit by integrating out nuisance parameters. An implementation of our method is freely available for download as a plugin to Cytoscape through our website and through Cytoscape itself.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

