In silico feasibility of novel biodegradation pathways for 1,2,4-trichlorobenzene
- PMID: 20122273
- PMCID: PMC2830930
- DOI: 10.1186/1752-0509-4-7
In silico feasibility of novel biodegradation pathways for 1,2,4-trichlorobenzene
Abstract
Background: Bioremediation offers a promising pollution treatment method in the reduction and elimination of man-made compounds in the environment. Computational tools to predict novel biodegradation pathways for pollutants allow one to explore the capabilities of microorganisms in cleaning up the environment. However, given the wealth of novel pathways obtained using these prediction methods, it is necessary to evaluate their relative feasibility, particularly within the context of the cellular environment.
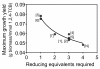
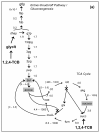
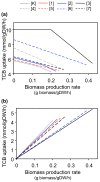
Results: We have utilized a computational framework called BNICE to generate novel biodegradation routes for 1,2,4-trichlorobenzene (1,2,4-TCB) and incorporated the pathways into a metabolic model for Pseudomonas putida. We studied the cellular feasibility of the pathways by applying metabolic flux analysis (MFA) and thermodynamic constraints. We found that the novel pathways generated by BNICE enabled the cell to produce more biomass than the known pathway. Evaluation of the flux distribution profiles revealed that several properties influenced biomass production: 1) reducing power required, 2) reactions required to generate biomass precursors, 3) oxygen utilization, and 4) thermodynamic topology of the pathway. Based on pathway analysis, MFA, and thermodynamic properties, we identified several promising pathways that can be engineered into a host organism to accomplish bioremediation.
Conclusions: This work was aimed at understanding how novel biodegradation pathways influence the existing metabolism of a host organism. We have identified attractive targets for metabolic engineers interested in constructing a microorganism that can be used for bioremediation. Through this work, computational tools are shown to be useful in the design and evaluation of novel xenobiotic biodegradation pathways, identifying cellularly feasible degradation routes.
Figures

References
-
- Jain RK, Kapur M, Labana S, Lal B, PM S, Bhattacharya D, Thakur IS. Microbial diversity: application of microorganisms for the biodegradation of xenobiotics. Curr Sci. 2005;89:101–112.
-
- Meer JR van der. In: Microbial biodegradation: Genomics and molecular biology. Diaz E, editor. Norfolk, UK: Caister Academic Press; 2008. A genomic view on the evolution of catabolic pathways and bacterial adaptation to xenobiotic compounds.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

