Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins
- PMID: 20122406
- PMCID: PMC6396293
- DOI: 10.1016/j.molcel.2009.12.030
Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins
Abstract
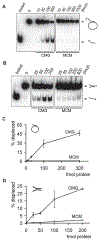
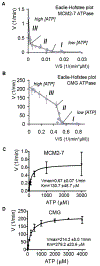
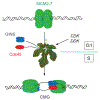
MCM2-7 proteins provide essential helicase functions in eukaryotes at chromosomal DNA replication forks. During the G1 phase of the cell cycle, they remain loaded on DNA but are inactive. We have used recombinant methods to show that the Drosophila MCM2-7 helicase is activated in complex with Cdc45 and the four GINS proteins (CMG complex). Biochemical activities of the MCM AAA+ motor are altered and enhanced through such associations: ATP hydrolysis rates are elevated by two orders of magnitude, helicase activity is robust on circular templates, and affinity for DNA substrates is improved. The GINS proteins contribute to DNA substrate affinity and bind specifically to the MCM4 subunit. All pairwise associations among GINS, MCMs, and Cdc45 were detected, but tight association takes place only in the CMG. The onset of DNA replication and unwinding may thus occur through allosteric changes in MCM2-7 affected by the association of these ancillary factors.
Copyright 2010 Elsevier Inc. All rights reserved.
Figures




References
-
- Aparicio OM, Weinstein DM, and Bell SP (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69. - PubMed
-
- Bell SP, and Dutta A (2002). DNA replication in eukaryotic cells. Annual Review of Biochemistry 71, 333–374. - PubMed
-
- Bochman ML, and Schwacha A (2007). Differences in the single-stranded DNA binding activities of MCM2-7 and MCM467: MCM2 and 5 define a slow ATP-dependent step. J. Biol. Chem 282, 33795–33804. - PubMed
-
- Bochman ML, and Schwacha A (2008). The MCM2-7 complex has in vitro helicase activity. Mol. Cell 31, 287–293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

