Bacterial chemoreceptors: providing enhanced features to two-component signaling
- PMID: 20122866
- PMCID: PMC2863001
- DOI: 10.1016/j.mib.2009.12.014
Bacterial chemoreceptors: providing enhanced features to two-component signaling
Abstract
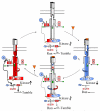
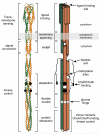
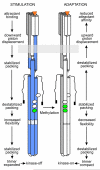
Bacteria perform chemotaxis utilizing core two-component signaling systems to which have been added enhanced features of signal amplification, sensory adaptation, molecular memory and high sensitivity over a wide dynamic range. Chemoreceptors are central to the enhancements. These transmembrane homodimers associate in trimers and in clusters of signaling complexes containing from a few to thousands of receptors. Receptor homodimers couple ligand occupancy and adaptational modification to transmembrane signaling. Trimers activate and control the histidine kinase. Clusters enable signal amplification, high sensitivity and adaptational assistance. Homodimer signaling initiates with helical piston sliding that is converted to modulation of competing packing modes of adjacent segments of an extended helical coiled coil. In trimers, signaling and coupling may involve switching between compact and expanded forms.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

