Multiple nucleotide preferences determine cleavage-site recognition by the HIV-1 and M-MuLV RNases H
- PMID: 20122939
- PMCID: PMC2830385
- DOI: 10.1016/j.jmb.2010.01.059
Multiple nucleotide preferences determine cleavage-site recognition by the HIV-1 and M-MuLV RNases H
Abstract
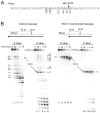
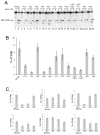
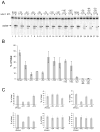
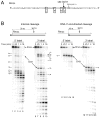
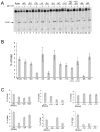
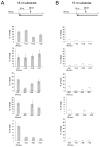
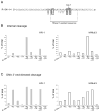
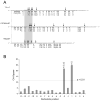
The RNase H activity of reverse transcriptase is required during retroviral replication and represents a potential target in antiviral drug therapies. Sequence features flanking a cleavage site influence the three types of retroviral RNase H activity: internal, DNA 3'-end-directed, and RNA 5'-end-directed. Using the reverse transcriptases of HIV-1 (human immunodeficiency virus type 1) and Moloney murine leukemia virus (M-MuLV), we evaluated how individual base preferences at a cleavage site direct retroviral RNase H specificity. Strong test cleavage sites (designated as between nucleotide positions -1 and +1) for the HIV-1 and M-MuLV enzymes were introduced into model hybrid substrates designed to assay internal or DNA 3'-end-directed cleavage, and base substitutions were tested at specific nucleotide positions. For internal cleavage, positions +1, -2, -4, -5, -10, and -14 for HIV-1 and positions +1, -2, -6, and -7 for M-MuLV significantly affected RNase H cleavage efficiency, while positions -7 and -12 for HIV-1 and positions -4, -9, and -11 for M-MuLV had more modest effects. DNA 3'-end-directed cleavage was influenced substantially by positions +1, -2, -4, and -5 for HIV-1 and positions +1, -2, -6, and -7 for M-MuLV. Cleavage-site distance from the recessed end did not affect sequence preferences for M-MuLV reverse transcriptase. Based on the identified sequence preferences, a cleavage site recognized by both HIV-1 and M-MuLV enzymes was introduced into a sequence that was otherwise resistant to RNase H. The isolated RNase H domain of M-MuLV reverse transcriptase retained sequence preferences at positions +1 and -2 despite prolific cleavage in the absence of the polymerase domain. The sequence preferences of retroviral RNase H likely reflect structural features in the substrate that favor cleavage and represent a novel specificity determinant to consider in drug design.
Copyright (c) 2010 Elsevier Ltd. All rights reserved.
Figures


References
-
- Arts EJ, LeGrice SFJ. Interaction of retroviral reverse transcriptase with template-primer duplexes during replication. Prog Nucleic Acid Res Mol Biol. 1998;58:339–393. - PubMed
-
- Telesnitsky A, Goff SP. Strong-stop Strand Transfer during Reverse Transcription. In: Skalka AM, Goff SP, editors. Reverse Transcriptase. Cold Spring Harbor Laboratory Press; Plainview, New York: 1993. pp. 49–83.
-
- Tisdale M, Schulze T, Larder BA, Moelling K. Mutations within the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase abolish virus infectivity. J Gen Virol. 1991;72 (Pt 1):59–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

