Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair
- PMID: 20122942
- PMCID: PMC2839085
- DOI: 10.1016/j.jmb.2010.01.049
Crystal structure of the first eubacterial Mre11 nuclease reveals novel features that may discriminate substrates during DNA repair
Abstract
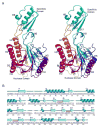
Mre11 nuclease plays a central role in the repair of cytotoxic and mutagenic DNA double-strand breaks. As X-ray structural information has been available only for the Pyrococcus furiosus enzyme (PfMre11), the conserved and variable features of this nuclease across the domains of life have not been experimentally defined. Our crystal structure and biochemical studies demonstrate that TM1635 from Thermotoga maritima, originally annotated as a putative nuclease, is an Mre11 endo/exonuclease (TmMre11) and the first such structure from eubacteria. TmMre11 and PfMre11 display similar overall structures, despite sequence identity in the twilight zone of only approximately 20%. However, they differ substantially in their DNA-specificity domains and in their dimeric organization. Residues in the nuclease domain are highly conserved, but those in the DNA-specificity domain are not. The structural differences likely affect how Mre11 from different organisms recognize and interact with single-stranded DNA, double-stranded DNA and DNA hairpin structures during DNA repair. The TmMre11 nuclease active site has no bound metal ions, but is conserved in sequence and structure with the exception of a histidine that is important in PfMre11 nuclease activity. Nevertheless, biochemical characterization confirms that TmMre11 possesses both endonuclease and exonuclease activities on single-stranded and double-stranded DNA substrates, respectively.
Copyright 2010 Elsevier Ltd. All rights reserved.
Figures



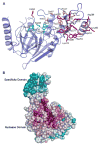


References
-
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. - PubMed
-
- Gellert M. Recent advances in understanding V(D)J recombination. Adv Immunol. 1997;64:39–64. - PubMed
-
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

