Animal models of mitochondrial DNA transactions in disease and ageing
- PMID: 20123011
- PMCID: PMC3320732
- DOI: 10.1016/j.exger.2010.01.019
Animal models of mitochondrial DNA transactions in disease and ageing
Abstract
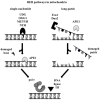
Mitochondrial DNA (mtDNA) transactions, processes that include mtDNA replication, repair, recombination and transcription constitute the initial stages of mitochondrial biogenesis, and are at the core of understanding mitochondrial biology and medicine. All of the protein players are encoded in nuclear genes: some are proteins with well-known functions in the nucleus, others are well-known mitochondrial proteins now ascribed new functions, and still others are newly discovered factors. In this article we review recent advances in the field of mtDNA transactions with a special focus on physiological studies. In particular, we consider the expression of variant proteins, or altered expression of factors involved in these processes in powerful model organisms, such as Drosophila melanogaster and the mouse, which have promoted recognition of the broad relevance of oxidative phosphorylation defects resulting from improper maintenance of mtDNA. Furthermore, the animal models recapitulate many phenotypes related to human ageing and a variety of different diseases, a feature that has enhanced our understanding of, and inspired theories about, the molecular mechanisms of such biological processes.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures


References
-
- Adán C, Matsushima Y, Hernández-Sierra R, Marco-Ferreres R, Fernández-Moreno MA, González-Vioque E, Calleja M, Aragón JJ, Kaguni LS, Garesse R. Mitochondrial transcription factor B2 is essential for metabolic function in Drosophila melanogaster development. J Biol Chem. 2008;283:12333–12342. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

